We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Karisma Moll/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | =Structure and Function of Dipeptidyl Peptidase IV ( | + | =Structure and Function of Dipeptidyl Peptidase IV (DPP-IV) in Humans= |
| - | <StructureSection load='6b1e' size='350' frame='true' side='right' caption='PDB 6B1E for | + | <StructureSection load='6b1e' size='350' frame='true' side='right' caption='PDB 6B1E for DPP-IV in complex with Vildagliptin' scene='10/1037489/Starter_dppiv_colors/1'> |
| Line 7: | Line 7: | ||
[https://www.sciencedirect.com/science/article/pii/S0304416520301483#t0005 Proline Peptidase Overview] | [https://www.sciencedirect.com/science/article/pii/S0304416520301483#t0005 Proline Peptidase Overview] | ||
=== History === | === History === | ||
| - | Dipeptidyl Peptidase IV's role in the inactivation of [https://en.wikipedia.org/wiki/Incretin incretin hormones] was discovered in the 1990s. Animal studies were conducted in the late 1990s, followed by human studies in the early 2000s. The first | + | Dipeptidyl Peptidase IV's role in the inactivation of [https://en.wikipedia.org/wiki/Incretin incretin hormones] was discovered in the 1990s. Animal studies were conducted in the late 1990s, followed by human studies in the early 2000s. The first DPP-IV inhibitors (sitagliptin, vildagliptin, alogliptin, saxagliptin, and linagliptin) were approved starting in 2006, and now serve as monotherapy or add-on to other therapies in a glucose-lowering capacity. <ref name="Ahrén">PMID:31275243</ref> Since their approval, there have been multiple long-term trials to continue exploring the long-term effects of these medications. It is known that gliptins directly impact the pancreas, kidney, heart, and vessels. The most investigated thus far is the effects of gliptins on renal and cardiovascular functioning. <ref name="Khalse">PMID:30294582</ref> Results of phase II and III trails indicated that gliptins did not harm the cardiovascular system. A meta-analysis implicated two possible beneficial effects for patients treated with these medications: a reduction in cardiovascular effects and a direct renoprotective effect. <ref name="Hocher">PMID:22947920</ref> |
=== Function === | === Function === | ||
| - | + | DPP-IV, a [https://en.wikipedia.org/wiki/Protein_moonlighting moonlighting protein], has been implicated in many functions and diseases of the body including glucose metabolism, cardiovascular disease, the stress response, autoimmune diseases (i.e. HIV/AIDS), inflammation, and tumor biology. <ref name="Zhong">PMID:26441982</ref> <ref name="Sharma">PMID:34411658</ref> <ref name="Huang">PMID:35309386</ref> The active site functions in two ways: it can bind inhibitors and it can truncate substrates. Inhibitors inhibit DPP-IV via [https://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibition], binding to the active site but are not degraded, so they remain bound and block the enzymatic activity. Substrates are truncated by DPP-IV by temporarily forming a covalent bond and ultimately being released in two pieces (the first two residues and the truncated protein). | |
== Structure == | == Structure == | ||
| - | Something about interactions... | ||
| - | <ref name="Hiramatsu">PMID:12646248</ref> | ||
=== Overview === | === Overview === | ||
| Line 27: | Line 25: | ||
<scene name='10/1037489/Sax_surface/3'>scene caption sax surface</scene> | <scene name='10/1037489/Sax_surface/3'>scene caption sax surface</scene> | ||
| - | + | DPP-IV is found in two forms in the body: a membrane bound monomer and a <scene name='10/1037489/Homodimer/1'>blood soluble homodimer</scene>. All structural renderings of DPP-IV start at the 39th residue, meaning it does not include the intracellular domain, transmembrane region, and part of the cleavage site. The monomer has 4 domains: {{font color|dimgray|DPP-IV cleavage stalk}}, {{font color|tomato|beta propeller}}, {{font color|khaki|cystine-rich region}}, and the {{font color|mediumseagreen|catalytic domain}}. | |
| - | The | + | The DPP-IV [https://en.wikipedia.org/wiki/Beta-propeller beta propeller] is notable as it differs from all the other enzymes in the [https://en.wikipedia.org/wiki/Dipeptidyl_peptidase dipeptidyl peptidase family]. In all other DPPs the beta propeller has ligand gating potential; however, the <scene name='10/1037489/Beta_propeller/2'>beta propeller</scene> is an asymmetrical 7 blade propeller that does not function as a ligand gate by rather acts as a binding site which allows DPP-IV to conjugate with [https://en.wikipedia.org/wiki/Adenosine_deaminase Adenosine Deaminase]. <ref name="Abbott">PMID:10583373</ref> |
The <scene name='10/1037489/Cystine_rich_region/1'>cystine rich region</scene> contains 6 cystine residues (C385, C394, C444, C447, C454, C472) that make <scene name='10/1037489/Disulfide_bonds/1'>three disulfide bonds</scene> that play a critical role in the tertiary structure and therefore function of the DPPIV enzyme. <ref name="Dobers">PMID:10931192</ref> | The <scene name='10/1037489/Cystine_rich_region/1'>cystine rich region</scene> contains 6 cystine residues (C385, C394, C444, C447, C454, C472) that make <scene name='10/1037489/Disulfide_bonds/1'>three disulfide bonds</scene> that play a critical role in the tertiary structure and therefore function of the DPPIV enzyme. <ref name="Dobers">PMID:10931192</ref> | ||
| - | The {{font color|mediumseagreen|catalytic domain}} is where | + | The {{font color|mediumseagreen|catalytic domain}} is where DPP-IV substrates are cleaved at he penultimate point or where inhibitors bind to prevent DPP-IV enzymatic activity.The <scene name='10/1037489/Catalytic_domain/1'>catalytic domain</scene> includes the {{font color|springgreen|S1 binding pocket}} and {{font color|teal|S2 binding pocket}}. The {{font color|springgreen|S1 binding pocket}} contains hydrophobic residues (W547, S630, Y631, V656, W659, Y662, Y666, N710, V711 and H740) that interact with either the substrate or inhibitor to keep it within the catalytic domain. The {{font color|teal|S2 binding pocket}} contains residues (E205, E206, Y662, S209, R358 and P357) that create hydrogen bonds with either substrate or inhibitor to additionally keep it in place to be cleaved by the catalytic triad. Within these <scene name='10/1037489/Bp_w_surface_new/1'>binding pockets</scene> is the <scene name='10/1037489/Catalytic_triad/8'>catalytic triad</scene>, assisted by the oxyanion hole (residue Y631). <ref name="Kim">PMID:30103438</ref> |
| - | <ref name="Hiramatsu"/> | + | <ref name="Hiramatsu">PMID:12646248</ref> |
=== Mechanism === | === Mechanism === | ||
<scene name='10/1037489/Glp1_peptide__scissile_bond/3'>scene caption scissile bond</scene> | <scene name='10/1037489/Glp1_peptide__scissile_bond/3'>scene caption scissile bond</scene> | ||
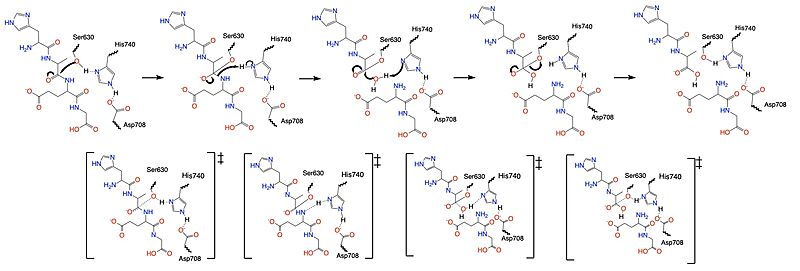
| - | Once a substrate is bound in the active site, | + | Once a substrate is bound in the active site, DPP-IV utilizes a [https://en.wikipedia.org/wiki/Enzyme_catalysis#Covalent_catalysis covalent catalysis] mechanism to cleave the substrate at the penultimate position. Asp708 of the <scene name='10/1037489/Catalytic_triad/7'>catalytic triad</scene> (Ser630, His 740, Asp708) pulls electron density from His740, allowing the histidine to pull electron density from Ser630, making serine a stronger nucleophile. The catalytic triad is assisted in this process by the oxyanion hole (residue Y631), providing stability and keeping the substrate in place. The water molecule attacks the carbonyl carbon, breaking the newly formed covalent bond, and releasing the first two residues of the starting substrate. The active site resets. |
| - | [[Image:DPPIV_Mech_SS.jpg|800 px|center|thumb|Figure 5. Mechanism for the cleavage of substrate of | + | [[Image:DPPIV_Mech_SS.jpg|800 px|center|thumb|Figure 5. Mechanism for the cleavage of substrate of DPP-IV via the catalytic triad.]] |
=== Inhibitors === | === Inhibitors === | ||
| - | Gliptins, a class of oral antidiabetic medications, are | + | Gliptins, a class of oral antidiabetic medications, are DPP-IV [https://en.wikipedia.org/wiki/Enzyme_inhibitor inhibitors]. The Food and Drug Administration (FDA) has approved the following: sitagliptin, alogliptin, saxagliptin, and linagliptin. The European Medicines Agency (EMA) has approved all of those aforementioned in addition to vildagliptin. Each gliptin is a [https://en.wikipedia.org/wiki/Small_molecule small molecule] (≤ 1000 Da). |
== Relevance == | == Relevance == | ||
| - | + | DPP-IV is known to cleave dozens of peptides including, but not limited to, regulatory peptides, neuropeptides, and chemokines. DPP-IV substrates are between 20 and 100 residues long, all of which contain a penultimate proline or alanine, indicating a stereochemical preference (though other penultimate residues are known to be cleaved but with reduced catalytic efficiency). | |
=== Diabetes === | === Diabetes === | ||
| - | <scene name='10/1037489/Glp1_peptide/1'>Glucagon-like Peptide-1</scene> (GLP-1) is a 30 amino acid long hormone which is secreted into the gut by intestinal epithelial L-cells.<ref name="Holst">PMID:17928588</ref> GLP-1 is secreted in response to meal intake and is responsible for stimulating insulin production. GLP-1 is a major peptide which regulates blood glucose levels and is also a major contributor to DPPIV substrates. The concentration of active GLP-1 is tightly regulated by DPPIV. GLP-1 is rapidly metabolized and degraded by DPPIV before even reaching the gut, which results in increased blood glucose levels. Insufficient GLP production or signaling in response to meal intake as been clinically associated with [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 diabetes] and morbidity given the close [https://en.wikipedia.org/wiki/Receptor_antagonist antagonist correlation] between GLP production and blood glucose levels. The antagonistic regulation of GLP-1 by | + | <scene name='10/1037489/Glp1_peptide/1'>Glucagon-like Peptide-1</scene> (GLP-1) is a 30 amino acid long hormone which is secreted into the gut by intestinal epithelial L-cells.<ref name="Holst">PMID:17928588</ref> GLP-1 is secreted in response to meal intake and is responsible for stimulating insulin production. GLP-1 is a major peptide which regulates blood glucose levels and is also a major contributor to DPPIV substrates. The concentration of active GLP-1 is tightly regulated by DPPIV. GLP-1 is rapidly metabolized and degraded by DPPIV before even reaching the gut, which results in increased blood glucose levels. Insufficient GLP production or signaling in response to meal intake as been clinically associated with [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 diabetes] and morbidity given the close [https://en.wikipedia.org/wiki/Receptor_antagonist antagonist correlation] between GLP production and blood glucose levels. The antagonistic regulation of GLP-1 by DPP-IV has made DPP-IV inhibition a very excellent candidate for pharmacological therapeutics. <ref name="Mulvihill">PMID:25216328</ref> |
=== HIV/AIDS === | === HIV/AIDS === | ||
Revision as of 13:56, 23 April 2024
Structure and Function of Dipeptidyl Peptidase IV (DPP-IV) in Humans
| |||||||||||
References
- ↑ Ahrén B. DPP-4 Inhibition and the Path to Clinical Proof. Front Endocrinol (Lausanne). 2019 Jun 19;10:376. PMID:31275243 doi:10.3389/fendo.2019.00376
- ↑ Khalse M, Bhargava A. A Review on Cardiovascular Outcome Studies of Dipeptidyl Peptidase-4 Inhibitors. Indian J Endocrinol Metab. 2018 Sep-Oct;22(5):689-695. PMID:30294582 doi:10.4103/ijem.IJEM_104_18
- ↑ Hocher B, Reichetzeder C, Alter ML. Renal and cardiac effects of DPP4 inhibitors--from preclinical development to clinical research. Kidney Blood Press Res. 2012;36(1):65-84. PMID:22947920 doi:10.1159/000339028
- ↑ Zhong J, Rajagopalan S. Dipeptidyl Peptidase-4 Regulation of SDF-1/CXCR4 Axis: Implications for Cardiovascular Disease. Front Immunol. 2015 Sep 25;6:477. PMID:26441982 doi:10.3389/fimmu.2015.00477
- ↑ Sharma A, Ren X, Zhang H, Pandey GN. Effect of depression and suicidal behavior on neuropeptide Y (NPY) and its receptors in the adult human brain: A postmortem study. Prog Neuropsychopharmacol Biol Psychiatry. 2022 Jan 10;112:110428. PMID:34411658 doi:10.1016/j.pnpbp.2021.110428
- ↑ Ntafam CN, Beutler BD, Harris RD. Incarcerated gravid uterus: A rare but potentially devastating obstetric complication. Radiol Case Rep. 2022 Mar 10;17(5):1583-1586. PMID:35309386 doi:10.1016/j.radcr.2022.02.034
- ↑ Abbott CA, McCaughan GW, Levy MT, Church WB, Gorrell MD. Binding to human dipeptidyl peptidase IV by adenosine deaminase and antibodies that inhibit ligand binding involves overlapping, discontinuous sites on a predicted beta propeller domain. Eur J Biochem. 1999 Dec;266(3):798-810. PMID:10583373 doi:10.1046/j.1432-1327.1999.00902.x
- ↑ Dobers J, Grams S, Reutter W, Fan H. Roles of cysteines in rat dipeptidyl peptidase IV/CD26 in processing and proteolytic activity. Eur J Biochem. 2000 Aug;267(16):5093-100. PMID:10931192 doi:10.1046/j.1432-1327.2000.01571.x
- ↑ Kim BR, Kim HY, Choi I, Kim JB, Jin CH, Han AR. DPP-IV Inhibitory Potentials of Flavonol Glycosides Isolated from the Seeds of Lens culinaris: In Vitro and Molecular Docking Analyses. Molecules. 2018 Aug 10;23(8):1998. PMID:30103438 doi:10.3390/molecules23081998
- ↑ Hiramatsu H, Kyono K, Higashiyama Y, Fukushima C, Shima H, Sugiyama S, Inaka K, Yamamoto A, Shimizu R. The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold. Biochem Biophys Res Commun. 2003 Mar 21;302(4):849-54. PMID:12646248
- ↑ Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev. 2007 Oct;87(4):1409-39. PMID:17928588 doi:10.1152/physrev.00034.2006
- ↑ Mulvihill EE, Drucker DJ. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr Rev. 2014 Dec;35(6):992-1019. PMID:25216328 doi:10.1210/er.2014-1035
Student Contributors
- Karisma Moll
- Merritt Jugo
- Sam Magnabosco