We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Proposed Function == | == Proposed Function == | ||
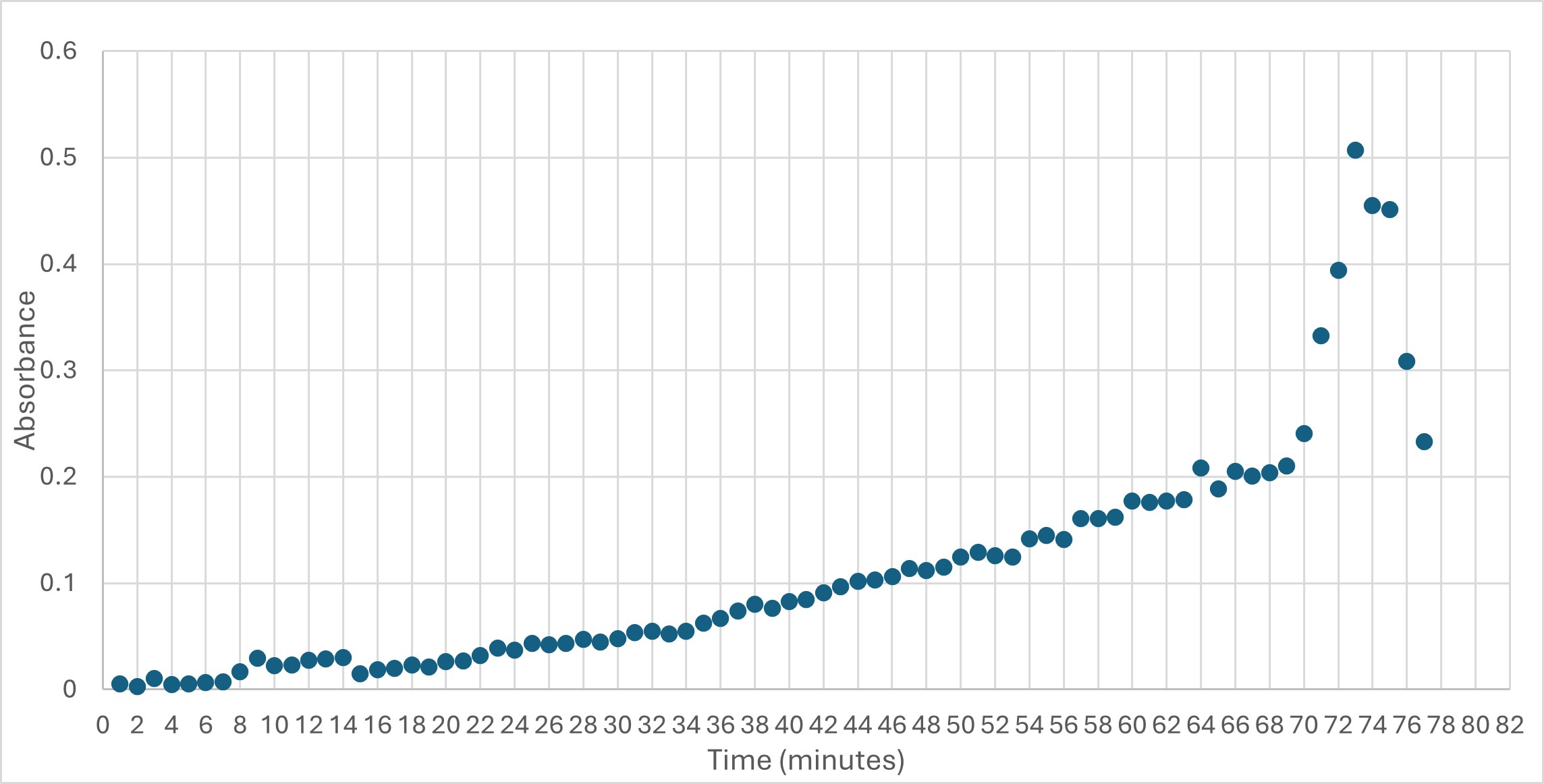
| - | This protein has alpha/beta-hydrolase activity and is of the esterase family. The SPRITE, BLAST, InterPro, and Dali Search along with other bioinformatics analyses consistently matched with carboxylesterases and other esterase-like proteins. SPRITE provided proteins that had similar residues in the active site of the enzyme. Multiple of the matching proteins were esterases and had an RMSD value of <0.5. The RMSD value gives insight into how similar the overlapping sections for the unknown and known proteins are. A value of less than 2 is desired when choosing proteins for reference. SwissDock and Chimera allowed for protein-ligand docking studies. Many of the highlighted interactions were ester-containing ligands and others were susceptible to hydrolysis. The isolated protein was introduced to 10 mM p-nitrophenyl acetate in a buffer with a pH of 6. Enzyme activity was measured, and the data was consistent with other hydrolases. | + | This protein has alpha/beta-hydrolase activity and is of the esterase family. The SPRITE, BLAST, InterPro, and Dali Search along with other bioinformatics analyses consistently matched with carboxylesterases and other esterase-like proteins. SPRITE provided proteins that had similar residues in the active site of the enzyme. Multiple of the matching proteins were esterases and had an RMSD value of <0.5. The RMSD value gives insight into how similar the overlapping sections for the unknown and known proteins are. A value of less than 2 is desired when choosing proteins for reference as a value closer to zero means less deviation between sites. SwissDock and Chimera allowed for protein-ligand docking studies. Many of the highlighted interactions were ester-containing ligands and others were susceptible to hydrolysis. The isolated protein was introduced to 10 mM p-nitrophenyl acetate in a buffer with a pH of 6. Enzyme activity was measured, and the data was consistent with other hydrolases. |
[[Image:Enzyme_Activity.jpeg]] | [[Image:Enzyme_Activity.jpeg]] | ||
== Structural highlights == | == Structural highlights == | ||
Revision as of 19:32, 24 April 2024
Protein 4DIU: Structure, Function, and Significance in Biological Systems
Think of this like an abstract. Include a few sentences about the most important data you found about what type of enzyme this is.
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644