We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Ben Whiteside
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
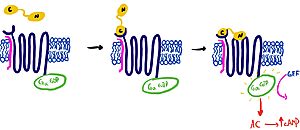
[https://en.wikipedia.org/wiki/Pramlintide Pramlintide] is a synthetic analog of amylin that is commonly used in accordance with mealtime [https://en.wikipedia.org/wiki/Insulin insulin] to help treat type 1 and 2 diabetic patients. This drug binds to AMYR competitively, increasing the AMYR GPCR signaling. Increased action of the AMYR receptor has been shown to modestly lower HbA1c levels, which is often accompanied by weight loss (cite 5). Pramlintide binds with more affinity than amylin due to mutations from hydrophobic residues A29, S28, S29, and S37 to proline. The proline residues increase the rigidity of the ligand by creating unfavorable phi and psi angles, which improves the ability of the ligand to bind AMYR. Pramlintide treatment has also been shown to consistently reduce [https://en.wikipedia.org/wiki/Amyloid_plaques Amyloid β plaque] aggregation in rodent models with [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer’s disease] (Gingell et al. 2014). | [https://en.wikipedia.org/wiki/Pramlintide Pramlintide] is a synthetic analog of amylin that is commonly used in accordance with mealtime [https://en.wikipedia.org/wiki/Insulin insulin] to help treat type 1 and 2 diabetic patients. This drug binds to AMYR competitively, increasing the AMYR GPCR signaling. Increased action of the AMYR receptor has been shown to modestly lower HbA1c levels, which is often accompanied by weight loss (cite 5). Pramlintide binds with more affinity than amylin due to mutations from hydrophobic residues A29, S28, S29, and S37 to proline. The proline residues increase the rigidity of the ligand by creating unfavorable phi and psi angles, which improves the ability of the ligand to bind AMYR. Pramlintide treatment has also been shown to consistently reduce [https://en.wikipedia.org/wiki/Amyloid_plaques Amyloid β plaque] aggregation in rodent models with [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer’s disease] (Gingell et al. 2014). | ||
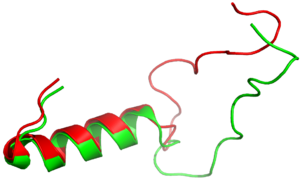
| - | It has been thought that [https://en.wikipedia.org/wiki/Missense_mutation missense mutations] | + | It has been thought that [https://en.wikipedia.org/wiki/Missense_mutation missense mutations] in residues C2 and C7 of the amylin peptide could lead to an increased risk of Alzheimer's Disease (CITE). Because of the rigidity these cysteine resides provide, reductions of their disulfide interaction leads to an increased risk of amyloid plaques due to amylin misfolding and forming aggregates. During drug design, pharmaceutical companies have focused on maintaining amylin residues C2 and C7, as well as K1, which forms a acts as a hydrogen bond donor for the <scene name='10/1038828/N_term_disulfidenew/1'>E294 Side Chain</scene> and main chain carbonyl. Additionally, pharmaceuticals companies have also opted to maintain residues <scene name='10/1038819/Amidated_c_term/9'>Y37 and T36</scene>, which are critical residues in stabilizing the C terminus of amylin to the receptor binding site. While there are hardly differences in the helical portion of amylin and the syntehtic analgoue pramlintide, there is a difference in the extended random coil at the C terminus. |
| + | |||
[[Image:align.png|300px|left|thumb|Figure 3:Amylin (green) aligned with Pramlintide (red)]] | [[Image:align.png|300px|left|thumb|Figure 3:Amylin (green) aligned with Pramlintide (red)]] | ||
| | ||
| Line 45: | Line 46: | ||
== Student Contributors == | == Student Contributors == | ||
| - | Andrew Helmerich,Mathias Vander Eide, Ben Whiteside | + | Andrew Helmerich, Mathias Vander Eide, Ben Whiteside |
Revision as of 20:52, 24 April 2024
AMYR
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev. 2015 Jul;67(3):564-600. PMID:26071095 doi:10.1124/pr.115.010629
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
Student Contributors
Andrew Helmerich, Mathias Vander Eide, Ben Whiteside