User:Jaelin Lunato/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
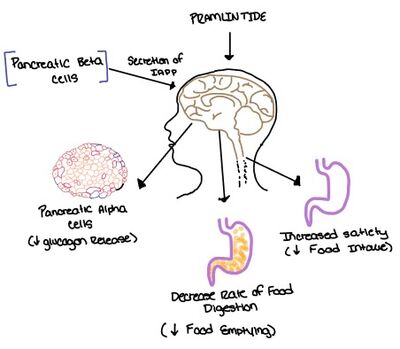
In the early 1900s, amyloid protein deposits were found in the pancreatic tissue of diabetic patients. Later in 1999, the structure of the [https://en.wikipedia.org/wiki/Calcitonin_receptor calcitonin receptors] were determined, and it was determined that the amylin receptor (AMYR)<ref name="7TYF">PMID:35324283</ref> had a core structure of the calcitonin receptor (CTR). Additionally, it was determined that AMYR had a [https://en.wikipedia.org/wiki/Receptor_activity-modifying_protein receptor activity-modifying protein] (RAMP) bound to the CTR core. In 2000, the CTR and a RAMP were co-expressed in a single cell, and later in 2016 the CTR and a RAMP were both found in rat brain cells. | In the early 1900s, amyloid protein deposits were found in the pancreatic tissue of diabetic patients. Later in 1999, the structure of the [https://en.wikipedia.org/wiki/Calcitonin_receptor calcitonin receptors] were determined, and it was determined that the amylin receptor (AMYR)<ref name="7TYF">PMID:35324283</ref> had a core structure of the calcitonin receptor (CTR). Additionally, it was determined that AMYR had a [https://en.wikipedia.org/wiki/Receptor_activity-modifying_protein receptor activity-modifying protein] (RAMP) bound to the CTR core. In 2000, the CTR and a RAMP were co-expressed in a single cell, and later in 2016 the CTR and a RAMP were both found in rat brain cells. | ||
| - | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-electron microscopy] (cryo-EM) was used to determine the structure of the calcitonin receptor and later the structure of AMYR. [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR] and [https://en.wikipedia.org/wiki/X-ray_crystallography x-ray crystallography] | + | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-electron microscopy] (cryo-EM) was used to determine the structure of the calcitonin receptor and later the structure of AMYR. [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR] and [https://en.wikipedia.org/wiki/X-ray_crystallography x-ray crystallography] were also used to determine the structure but the resolution was poor relative to cryo-EM. |
== Structure == | == Structure == | ||
=== Cellular Domains === | === Cellular Domains === | ||
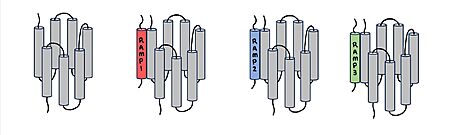
| - | [[Image:RAMP_diagram.jpg|450px|right|thumb|'''Figure 1.''' Three different RAMPs to compose AMY1R, AMY2R, and AMY3R when associated with the calcitonin receptor (shown in grey). RAMP1 is | + | [[Image:RAMP_diagram.jpg|450px|right|thumb|'''Figure 1.''' Three different RAMPs to compose AMY1R, AMY2R, and AMY3R when associated with the calcitonin receptor (shown in grey). RAMP1 is red, RAMP2 is blue, and RAMP3 is green.]] AMYR has three domains: extracellular, transmembrane, and intracellular. The CTR and the RAMP have both <scene name='10/1037495/Extracellular_domain/7'>extracellular domains</scene> and <scene name='10/1037495/Transmembrane/6'>transmembrane domains</scene>. The calcitonin receptor is a 7-pass helix chain to which RAMP binds to become the amylin receptor.<ref name="Hay">PMID:26071095</ref> The ligand, amylin, binds within the transmembrane domain. The [https://en.wikipedia.org/wiki/G_protein G protein] is located <scene name='10/1037495/Intracellular_domain/2'>inside the cell</scene>. The location of each component of the amylin receptor is essential in determining structure and function. For example, it is important for the G protein to be located inside the cell in order to initiate a signal cascade for cell signaling.<ref name="Cao">PMID:35324283</ref> |
=== CTR and RAMP Heterodimer === | === CTR and RAMP Heterodimer === | ||
Revision as of 01:01, 26 April 2024
Amylin Receptor (AMYR)
| |||||||||||
References
- ↑ Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
- ↑ 2.0 2.1 2.2 2.3 2.4 Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev. 2015 Jul;67(3):564-600. PMID:26071095 doi:10.1124/pr.115.010629
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
- ↑ 4.0 4.1 Press, M., Jung, T., Konig, J., Grune, T., & Hohn, A. (2019) Protein aggregates and proteostasis in aging: Amylin and β-cell function. Mechanisms of Ageing and Development. 3, 46-54. DOI:10.1016/j.mad.2018.03.010
- ↑ Mathiesen DS, Lund A, Vilsbøll T, Knop FK, Bagger JI. Amylin and Calcitonin: Potential Therapeutic Strategies to Reduce Body Weight and Liver Fat. Front Endocrinol (Lausanne). 2021 Jan 8;11:617400. PMID:33488526 doi:10.3389/fendo.2020.617400
- ↑ Thapa, G., Kumari, A., Dasgupta, D., Bandyopadhy, S., Sarkar, N., Roy, K., Karunakaran, G., Kazmi, I., Karmakar, S., & Chakraborty, M. (2023). Chapter 5- Insight into the mechanism of action of anti-diabetic drugs. How Synthetic Drugs Work. 95-122. DOI:10.1016/B978-0-323-99855-0.00005-1
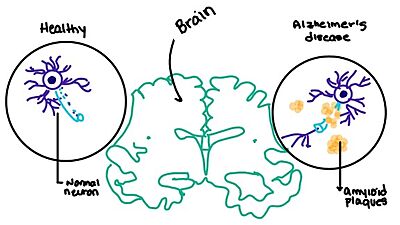
- ↑ Grizzanti J, Corrigan R, Casadesus G. Neuroprotective Effects of Amylin Analogues on Alzheimer's Disease Pathogenesis and Cognition. J Alzheimers Dis. 2018;66(1):11-23. PMID:30282360 doi:10.3233/JAD-180433