User:Chloe Tucker/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
This is a sample scene created with SAT to of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
=== Binding/Active Site of GIPR with GIP === | === Binding/Active Site of GIPR with GIP === | ||
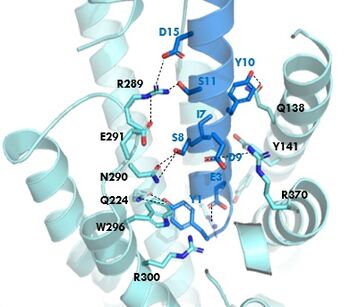
| - | [[Image:GIP_hydrogen_bonds.jpg|350 px|right|thumb|Figure 1. GIPR and GIP residue interactions]] | + | [[Image:GIP_hydrogen_bonds.jpg|350 px|right|thumb|Figure 1. GIPR (light blue) and GIP (dark blue) residue interactions]] |
The <scene name='10/1038815/Overview/7'>binding site</scene> of GIP with the GIP receptor (GIPR) is where the N-term of GIP binds with the transmembrane domain of the GIPR. The first interaction formed with GIPR and the N-term of GIP is a hydrogen bond between Tyrosine 1 (Y1) and Glutamine 224 (Q224) to activate the G-protein to start sending signals to the cell. | The <scene name='10/1038815/Overview/7'>binding site</scene> of GIP with the GIP receptor (GIPR) is where the N-term of GIP binds with the transmembrane domain of the GIPR. The first interaction formed with GIPR and the N-term of GIP is a hydrogen bond between Tyrosine 1 (Y1) and Glutamine 224 (Q224) to activate the G-protein to start sending signals to the cell. | ||
Many other <scene name='10/1038815/Active_site/3'>residues</scene> within the binding site are forming hydrogen bonds and hydrophobic interactions between the ligand and the receptor. The N-term binds more strongly than the C-term and there are many different residues contributing to this, including, Tyrosine 1 (Y1) and Tryptophan (W296) are forming aromatic interactions. Serine 8 (S8) and Asparagine 290 (N290) are forming two hydrogen bonds with each other. Aspartate 9 (D9) is forming another hydrogen bond with Arginine 370 (R370)<ref name="Sun"/>. '''Figure 1''' shows many of the interactions between GIP and GIPR. All of these interactions are responsible for the binding affinity, the strength of the attraction, between the two proteins. These hydrogen bonds lead to the activation of cell signaling and when this binding is somehow disrupted, that is what causes different diseases like diabetes. | Many other <scene name='10/1038815/Active_site/3'>residues</scene> within the binding site are forming hydrogen bonds and hydrophobic interactions between the ligand and the receptor. The N-term binds more strongly than the C-term and there are many different residues contributing to this, including, Tyrosine 1 (Y1) and Tryptophan (W296) are forming aromatic interactions. Serine 8 (S8) and Asparagine 290 (N290) are forming two hydrogen bonds with each other. Aspartate 9 (D9) is forming another hydrogen bond with Arginine 370 (R370)<ref name="Sun"/>. '''Figure 1''' shows many of the interactions between GIP and GIPR. All of these interactions are responsible for the binding affinity, the strength of the attraction, between the two proteins. These hydrogen bonds lead to the activation of cell signaling and when this binding is somehow disrupted, that is what causes different diseases like diabetes. | ||
=== Binding/Active Site of GIPR with Tirzepatide === | === Binding/Active Site of GIPR with Tirzepatide === | ||
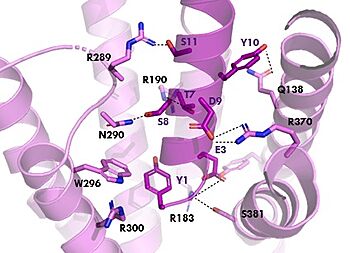
| - | [[Image:TZ_hydrogen_bonds.jpg|350 px|right|thumb|Figure 2. | + | [[Image:TZ_hydrogen_bonds.jpg|350 px|right|thumb|Figure 2. GIPR (pink) and Tirzepatide (dark pink) residue interactions]] |
The <scene name='10/1038815/Tirzepatide_active_site/4'>binding site</scene> with Tirzepatide is the same as GIP with the N-term binding to the transmembrane domain and activating cellular signaling.The <scene name='10/1038815/Tirzepatide_residues/7'>residues</scene> are fairly similar just in a different conformations, which is allowing for more hydrogen bonding. '''Figure 2''' shows the interactions between GIPR and Tirzepatide, along with the slight changes in conformation and hydrogen bonding. In Tirzepatide, the most noticeable conformational change is the Tyrosine 1 (Y1) residue. It is now facing up towards Arginine 190 (R190) where they can now form a new hydrogen bond that was not present in GIP. The more hydrogen bonding there is between molecules, the stronger the binding between the two molecules will be. This can lead to the conclusion that GIPR has a higher binding affinity for Tirzepatide than GIP itself<ref name="Sun"/>. | The <scene name='10/1038815/Tirzepatide_active_site/4'>binding site</scene> with Tirzepatide is the same as GIP with the N-term binding to the transmembrane domain and activating cellular signaling.The <scene name='10/1038815/Tirzepatide_residues/7'>residues</scene> are fairly similar just in a different conformations, which is allowing for more hydrogen bonding. '''Figure 2''' shows the interactions between GIPR and Tirzepatide, along with the slight changes in conformation and hydrogen bonding. In Tirzepatide, the most noticeable conformational change is the Tyrosine 1 (Y1) residue. It is now facing up towards Arginine 190 (R190) where they can now form a new hydrogen bond that was not present in GIP. The more hydrogen bonding there is between molecules, the stronger the binding between the two molecules will be. This can lead to the conclusion that GIPR has a higher binding affinity for Tirzepatide than GIP itself<ref name="Sun"/>. | ||
=== Isoleucine vs. Threonine === | === Isoleucine vs. Threonine === | ||
Revision as of 20:35, 26 April 2024
GIP and GIP-R
| |||||||||||
References
Student Contributors
- Chloe Tucker
- Mandy Bechman