We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='4DIU' size='340' side='right' caption='Structural Model of Protein 4DIU' scene=''> | ||
==Protein 4DIU: Structure, Function, and Significance in Biological Systems== | ==Protein 4DIU: Structure, Function, and Significance in Biological Systems== | ||
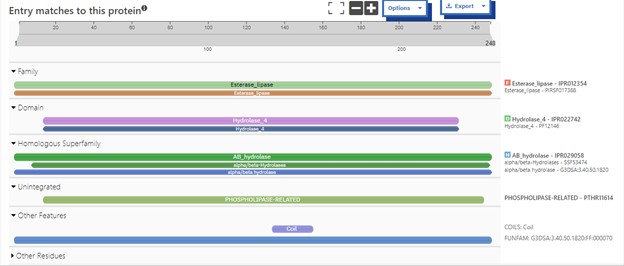
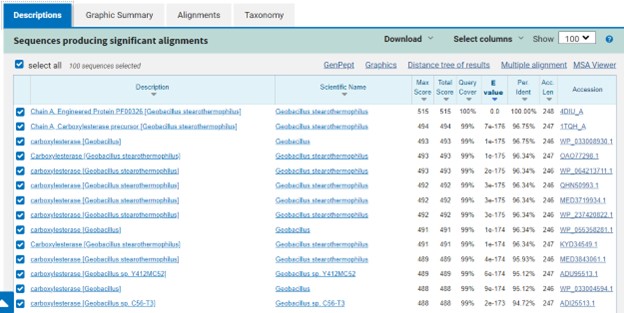
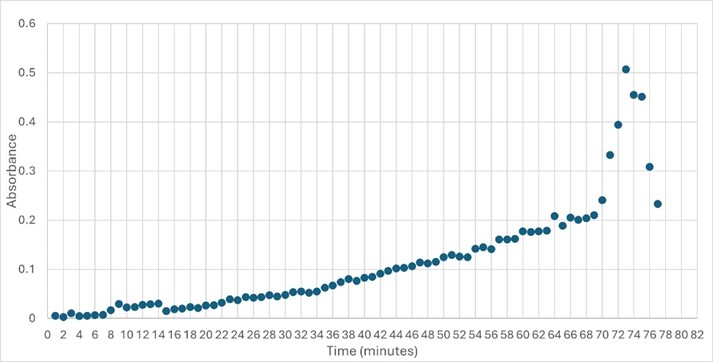
This study was designed to determine the function of unknown protein 4DIU and its optimal conditions. The protein 4DIU has been identified and characterized as a potential esterase enzyme based on in-depth analyses including bioinformatics, molecular docking studies, and laboratory experiments. Online tools such as BLAST, Dali, SPRITE, and InterPro often matched 4DIU with carboxylesterases and other esterase-like proteins. This data suggests that its function involves hydrolysis of esters. In part to data from Swiss Dock, 4DIU is predicted to have binding sites with an affinity to substrates such as acetate, butyrate, phosphate, proline, decanoate, and dodecanoate. The protein 4DIU was grown, harvested, and isolated in a laboratory. A Bradford assay was completed to determine the concentration of protein in each elution from a column. SDS-PAGE analysis was used to confirm the presence and identity of the protein via molecular weight. Once identified the enzyme's activity on the substrate p-nitrophenyl acetate was measured. This substrate was chosen because many hydrolases have an affinity for it and can hydrolyze the substrate. It was determined that the 4DIU protein functions best at a pH of 6. Using the change in absorbance and Beer's Law the amount of units of protein was calculated. | This study was designed to determine the function of unknown protein 4DIU and its optimal conditions. The protein 4DIU has been identified and characterized as a potential esterase enzyme based on in-depth analyses including bioinformatics, molecular docking studies, and laboratory experiments. Online tools such as BLAST, Dali, SPRITE, and InterPro often matched 4DIU with carboxylesterases and other esterase-like proteins. This data suggests that its function involves hydrolysis of esters. In part to data from Swiss Dock, 4DIU is predicted to have binding sites with an affinity to substrates such as acetate, butyrate, phosphate, proline, decanoate, and dodecanoate. The protein 4DIU was grown, harvested, and isolated in a laboratory. A Bradford assay was completed to determine the concentration of protein in each elution from a column. SDS-PAGE analysis was used to confirm the presence and identity of the protein via molecular weight. Once identified the enzyme's activity on the substrate p-nitrophenyl acetate was measured. This substrate was chosen because many hydrolases have an affinity for it and can hydrolyze the substrate. It was determined that the 4DIU protein functions best at a pH of 6. Using the change in absorbance and Beer's Law the amount of units of protein was calculated. | ||
| - | + | ||
<ref>PMID:21638687</ref> | <ref>PMID:21638687</ref> | ||
== Materials == | == Materials == | ||
Revision as of 23:56, 27 April 2024
| |||||||||||
References
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the Para-Nitrophenol Catabolic Pathway, and Characterization of Three Enzymes Involved in the Hydroquinone Pathway, in Pseudomonas Sp. 1-7. BMC Microbiology 2012, 12 (1). https://doi.org/10.1186/1471-2180-12-27.
- ↑ Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346.