Protein 4DIU: Structure, Function, and Significance in Biological Systems
This study was designed to determine the function of unknown protein 4DIU and its optimal conditions. The protein 4DIU has been identified and characterized as a potential esterase enzyme based on in-depth analyses including bioinformatics, molecular docking studies, and laboratory experiments. Online tools such as BLAST, Dali, SPRITE, and InterPro often matched 4DIU with carboxylesterases and other esterase-like proteins. This data suggests that its function involves hydrolysis of esters. Results from Swiss Dock and Chimera predict protein 4DIU to have binding sites with an affinity to substrates such as acetate, butyrate, phosphate, proline, decanoate, and dodecanoate. The protein 4DIU was grown, harvested, and isolated in a laboratory. A Bradford assay was completed to determine the concentration of protein in each elution from a column. SDS-PAGE analysis was used to confirm the presence and identity of the protein via molecular weight. Once identified the enzyme's activity on the substrate p-nitrophenyl acetate was measured. This substrate was chosen because many hydrolases have an affinity for it and can hydrolyze the substrate. It was determined that the 4DIU protein functions best at a pH of 6. Using the change in absorbance and Beer's Law the amount of units of protein was calculated.
Materials
- Buffers: Sodium Phosphate buffer, Cell Lysis Buffer Tris-HCl, 10X SDS-PAGE Buffer, Re-Suspension Buffer, 1X Wash Buffer, 1X Elution Buffer
- Solutions for SDS-Page: Coomassie Blue Stain, and Destain
- Pre-cast SDS-Page Gel
- E. Coli
Overview of Esterase
The role in the body this would play based on results. Any potential implications of the enzyme if disrupted based on findings and related enzymes.
Proposed Function
This protein has alpha/beta-hydrolase activity and is of the esterase family. The SPRITE, BLAST, InterPro, and Dali Search along with other bioinformatics analyses consistently matched with carboxylesterases and other esterase-like proteins. SPRITE provided proteins that had similar residues in the active site of the enzyme. Multiple of the matching proteins were esterases and had an RMSD value of <0.5. The RMSD value gives insight into how similar the overlapping sections for the unknown and known proteins are. A value of less than 2 is desired when choosing proteins for reference as a value closer to zero means less deviation between sites.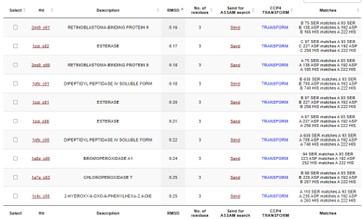
Additional evidence is provided by InteroPro which provided more information about the classification of the protein. As well, the BLAST search gave results of matching proteins and the specific functions of the proteins. All results again matched what was suggested by Sprite and Blast, further solidifying that 4DIU is an esterase with alpha/beta-hydrolase activity.
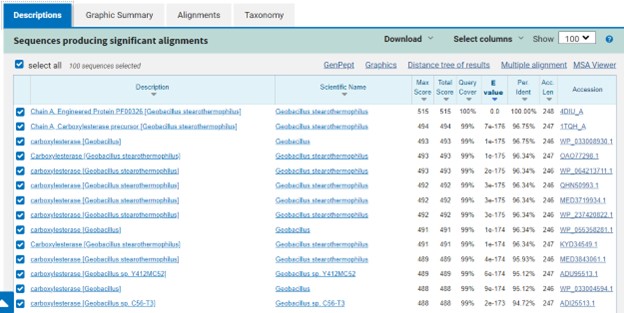
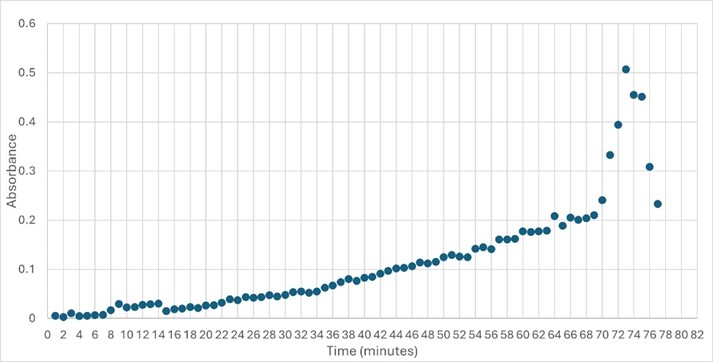
SwissDock and Chimera allowed for protein-ligand docking studies. Many of the highlighted interactions were ester-containing ligands and others were susceptible to hydrolysis. The hydrolase activity of the protein needed to be tested with a substrate known to work with hydrolases. This substrate is p-nitrophenyl acetate PNP when hydrolyzed produces a yellow colored product. The yellow product will lead to the solution absorbing more at 405nm. [1]. This allows for the activity of the enzyme to be tracked by measuring the increase in absorbance over time. The isolated protein was introduced to p-nitrophenyl acetate in a buffer with a determined optimal pH of 6. Enzyme activity was measured via a change in absorbance at 405nm over 80 minutes. The data collected was consistent with that of hydrolases in other studies.[2]
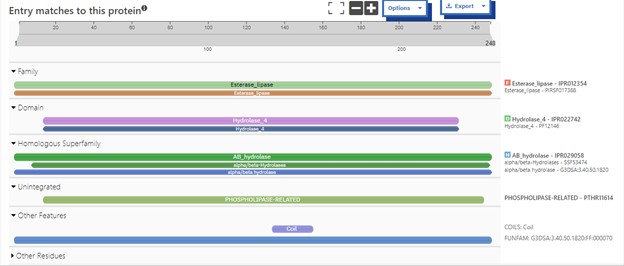
Structural highlights of 4DIU
[3]
Highlight the data that helped you come to your conclusion here including any relevant figures. Make sure to include potential substrates and binding sites.
Conclusions
Overall, the research question was answered and the hypothesis was correct. Protein 4DIU has been identified as belonging to the esterase family and has hydrolytic activity. It appears that the optimal pH for this enzyme is 6 and it is active with p-nitrophenyl acetate. These results are fairly accurate and precise as two trials were run at a pH of 6 and the same trend was seen. In future experiments it would be beneficial to test the activity at lower pHs such as 3, 4, and 5. Additionally, test some of the substrates that were molded to work to see if the bioinformatic data is trustworthy.



