User:Camille Gaudet/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
=== Tirzepatide Structure=== | === Tirzepatide Structure=== | ||
| - | Structurally, as a dual agonist, <scene name='10/1037488/Tirz-bound/3'>Tirzepatide</scene> closely resembles GIP and GLP-1. The sequence alignment of all three polypeptides showcases the highly integrated nature of both GIP and GLP-1 into the amino acid sequence of Tirzepatide | + | Structurally, as a dual agonist, <scene name='10/1037488/Tirz-bound/3'>Tirzepatide</scene> closely resembles GIP and GLP-1. The sequence alignment of all three polypeptides showcases the highly integrated nature of both GIP and GLP-1 into the amino acid sequence of Tirzepatide with very few alterations (Figure 2). [[Image:GIPGLPTirz.png|700 px|right|thumb|Figure 2. Sequence alignment of GLP-1, GIP, and Tirzepatide. Residues shown in black are found in all three amino acid sequences. Pink or red coloration denotes residues that are unique to GIP or GLP-1, respectively. The sequence of tirzepatide is colored accordingly, and residues differing from both GIP and GLP-1 are highlighted in blue. Residues that differ across all three structures are boxed.]] The <scene name='10/1037488/Gip-y1-intrxn-still/8'>conserved Tyr1</scene> residue allows for simulation of a highly similar interaction between Tirzepatide and the GIP receptor. Had this been altered, Tirzepatide would not nearly bind with as high of an affinity for GIPR. Several <scene name='10/1037488/Tirz-bound-differences/3'>unique residues in Tirzepatide</scene> do not align with either the sequence of GIP or GLP-1 and are located toward the C-terminus end of the structure. They include Ala21, Gln24, Ile27, and Gly30. The <scene name='10/1037488/Gip-differences/6'>corresponding GIP residues</scene> are Asp21, Asn24, Leu27, and Lys30. Interestingly, these residues do not interact with the receptor, thus their mutation doesn't alter the overall binding affinity of Tirzepatide to GIPR. |
Revision as of 21:46, 28 April 2024
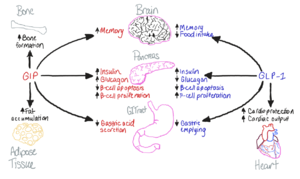
Glucose-dependent Insulinotropic Polypeptide Receptor
| |||||||||||
References
- ↑ 1.0 1.1 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
Student Contributors
- Camille Gaudet
- Sara Kalkhoff