We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
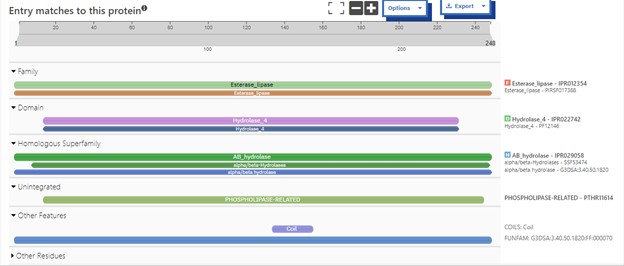
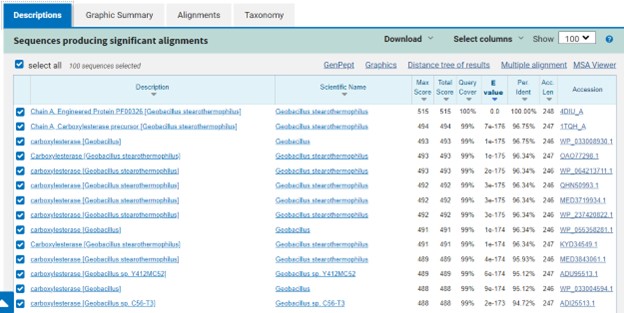
Additional evidence is provided by InteroPro which provided more information about the classification of the protein. As well, the BLAST search gave results of matching proteins and the specific functions of the proteins. All results again matched what was suggested by Sprite and Blast, further solidifying that 4DIU is an esterase with alpha/beta-hydrolase activity. | Additional evidence is provided by InteroPro which provided more information about the classification of the protein. As well, the BLAST search gave results of matching proteins and the specific functions of the proteins. All results again matched what was suggested by Sprite and Blast, further solidifying that 4DIU is an esterase with alpha/beta-hydrolase activity. | ||
[[Image:InterPro.jpg]] | [[Image:InterPro.jpg]] | ||
| + | |||
'''Figure 2: '''Interpro Scan results for 4DIU displaying proposed characterization | '''Figure 2: '''Interpro Scan results for 4DIU displaying proposed characterization | ||
[[Image:Blast.jpg]] | [[Image:Blast.jpg]] | ||
| + | |||
'''Figure 3: '''Blast search for protein sequences matching 4DIU | '''Figure 3: '''Blast search for protein sequences matching 4DIU | ||
| Line 33: | Line 35: | ||
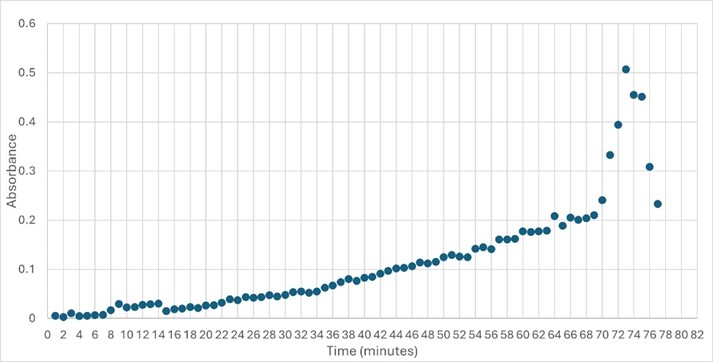
</ref>. This allows for the activity of the enzyme to be tracked by measuring the increase in absorbance over time. The isolated protein was introduced to p-nitrophenyl acetate in a buffer with a determined optimal pH of 6. Enzyme activity was measured via a change in absorbance at 405nm over 80 minutes. The data collected was consistent with that of hydrolases in other studies.<ref>Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346. | </ref>. This allows for the activity of the enzyme to be tracked by measuring the increase in absorbance over time. The isolated protein was introduced to p-nitrophenyl acetate in a buffer with a determined optimal pH of 6. Enzyme activity was measured via a change in absorbance at 405nm over 80 minutes. The data collected was consistent with that of hydrolases in other studies.<ref>Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346. | ||
</ref> | </ref> | ||
| + | |||
| + | '''Figure 3: ''' Trial 1 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 8 buffer | ||
| + | |||
| + | '''Figure 4: ''' Trial 1 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | ||
[[Image:Enzymeactivity.jpeg]] | [[Image:Enzymeactivity.jpeg]] | ||
| - | '''Figure | + | |
| + | '''Figure 5: ''' Trial 2 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | ||
== Structural highlights of 4DIU == | == Structural highlights of 4DIU == | ||
Revision as of 23:54, 28 April 2024
| |||||||||||
References
- ↑ Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the Para-Nitrophenol Catabolic Pathway, and Characterization of Three Enzymes Involved in the Hydroquinone Pathway, in Pseudomonas Sp. 1-7. BMC Microbiology 2012, 12 (1). https://doi.org/10.1186/1471-2180-12-27.
- ↑ Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346.
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644