User:Camille Gaudet/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 30: | Line 30: | ||
=== Tirzepatide Structure=== | === Tirzepatide Structure=== | ||
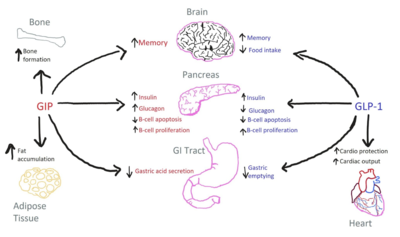
Structurally, as a dual agonist, <scene name='10/1037488/Tirz-bound/3'>Tirzepatide</scene> closely resembles GIP and GLP-1. The sequence alignment of all three polypeptides showcases the highly integrated nature of both GIP and GLP-1 into the amino acid sequence of Tirzepatide with very few alterations (Figure 2). [[Image:GIPGLPTirz.png|700 px|right|thumb|Figure 2. Sequence alignment of GLP-1, GIP, and Tirzepatide. Residues shown in black are found in all three amino acid sequences. Pink or red coloration denotes residues that are unique to GIP or GLP-1, respectively. The sequence of tirzepatide is colored accordingly, and residues differing from both GIP and GLP-1 are highlighted in blue. Residues that differ across all three structures are boxed.]] The <scene name='10/1037488/Tirz-y1conserved/2'>conserved Tyr1 residue</scene> allows for simulation of a highly similar interaction between Tirzepatide and the GIP receptor. Had this been altered, Tirzepatide would not nearly bind with as high of an affinity for GIPR. The Tyr1 residue facilitates strong contacts with the TMD core. Several <scene name='10/1037488/Tirz-bound-differences/3'>unique residues in Tirzepatide</scene> do not align with either the sequence of GIP or GLP-1 and are located toward the C-terminus end of the structure. They include Ala21, Gln24, Ile27, and Gly30. The <scene name='10/1037488/Gip-differences/6'>corresponding GIP residues</scene> are Asp21, Asn24, Leu27, and Lys30. Interestingly, these residues do not interact with the receptor, thus their mutation doesn't alter the overall binding affinity of Tirzepatide to GIPR.<ref name="Sun">PMID: 35333651</ref> Other significant structural modifications were made to maximize GIPR-Tirz interactions.The AIB (alpha-aminoisobutyric) residues (Figure 2) prevent degradation of the polypeptide by peptidases such as DPP-4.<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> | Structurally, as a dual agonist, <scene name='10/1037488/Tirz-bound/3'>Tirzepatide</scene> closely resembles GIP and GLP-1. The sequence alignment of all three polypeptides showcases the highly integrated nature of both GIP and GLP-1 into the amino acid sequence of Tirzepatide with very few alterations (Figure 2). [[Image:GIPGLPTirz.png|700 px|right|thumb|Figure 2. Sequence alignment of GLP-1, GIP, and Tirzepatide. Residues shown in black are found in all three amino acid sequences. Pink or red coloration denotes residues that are unique to GIP or GLP-1, respectively. The sequence of tirzepatide is colored accordingly, and residues differing from both GIP and GLP-1 are highlighted in blue. Residues that differ across all three structures are boxed.]] The <scene name='10/1037488/Tirz-y1conserved/2'>conserved Tyr1 residue</scene> allows for simulation of a highly similar interaction between Tirzepatide and the GIP receptor. Had this been altered, Tirzepatide would not nearly bind with as high of an affinity for GIPR. The Tyr1 residue facilitates strong contacts with the TMD core. Several <scene name='10/1037488/Tirz-bound-differences/3'>unique residues in Tirzepatide</scene> do not align with either the sequence of GIP or GLP-1 and are located toward the C-terminus end of the structure. They include Ala21, Gln24, Ile27, and Gly30. The <scene name='10/1037488/Gip-differences/6'>corresponding GIP residues</scene> are Asp21, Asn24, Leu27, and Lys30. Interestingly, these residues do not interact with the receptor, thus their mutation doesn't alter the overall binding affinity of Tirzepatide to GIPR.<ref name="Sun">PMID: 35333651</ref> Other significant structural modifications were made to maximize GIPR-Tirz interactions.The AIB (alpha-aminoisobutyric) residues (Figure 2) prevent degradation of the polypeptide by peptidases such as DPP-4.<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> | ||
| - | Because Tirzepatide was modeled after two distinct polypeptides, its binding to each of the receptors has hallmark differences. The Tirz-GIPR complex is rotated ~8.3º compared to Tirz-GLP-1R, with the C-terminus translated closer to the TMD core. ECL1 interactions with Tirzepatide are diminished in GIPR due to the presence of several repeating proline residues (P195, P197, and P199). Fortunately, the ɑ-helical extension provides another recognition point for Tirzepatide; a hydrogen bond between <scene name='10/1037488/Tirz-y10/1'>Y10 (Tirz) and Q138 (GIPR)</scene> and stacking between K16 (Tirz) and F127 (GIPR). Primary interactions between the receptor and Tirzepatide occur within the first 27 residues (Figure 3).<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref>[[Image:GIPRTirz-binding.png|700 px|right|thumb|Figure 3. Amino acid sequence of Tirzepatide and the type of interaction it forms with the GIP receptor. Salt bridges (blue), hydrogen bonds (red), pi stacking (orange), and Van der Waals (gray) interactions are highlighted.]] | + | Because Tirzepatide was modeled after two distinct polypeptides, its binding to each of the receptors has hallmark differences. The Tirz-GIPR complex is rotated ~8.3º compared to Tirz-GLP-1R, with the C-terminus translated closer to the TMD core. ECL1 interactions with Tirzepatide are diminished in GIPR due to the presence of several <scene name='10/1037488/Gip-r_receptor-centered/7'>repeating proline residues (P195, P197, and P199)</scene>. Fortunately, the ɑ-helical extension provides another recognition point for Tirzepatide; a hydrogen bond between <scene name='10/1037488/Tirz-y10/1'>Y10 (Tirz) and Q138 (GIPR)</scene> and stacking between K16 (Tirz) and F127 (GIPR). Primary interactions between the receptor and Tirzepatide occur within the first 27 residues (Figure 3).<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> |
| + | [[Image:GIPRTirz-binding.png|700 px|right|thumb|Figure 3. Amino acid sequence of Tirzepatide and the type of interaction it forms with the GIP receptor. Salt bridges (blue), hydrogen bonds (red), pi stacking (orange), and Van der Waals (gray) interactions are highlighted.]] | ||
| + | |||
Revision as of 02:37, 29 April 2024
Glucose-dependent Insulinotropic Polypeptide Receptor
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ 2.0 2.1 2.2 2.3 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
- ↑ 3.0 3.1 Chavda VP, Ajabiya J, Teli D, Bojarska J, Apostolopoulos V. Tirzepatide, a New Era of Dual-Targeted Treatment for Diabetes and Obesity: A Mini-Review. Molecules. 2022 Jul 5;27(13):4315. PMID:35807558 doi:10.3390/molecules27134315
- ↑ 4.0 4.1 4.2 Zhao F, Zhou Q, Cong Z, Hang K, Zou X, Zhang C, Chen Y, Dai A, Liang A, Ming Q, Wang M, Chen LN, Xu P, Chang R, Feng W, Xia T, Zhang Y, Wu B, Yang D, Zhao L, Xu HE, Wang MW. Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Nat Commun. 2022 Feb 25;13(1):1057. PMID:35217653 doi:10.1038/s41467-022-28683-0
Student Contributors
- Camille Gaudet
- Sara Kalkhoff