The Ohr protein (organic hydroperoxide resistance) is a thiol-dependent Cys-based peroxidase exclusive to bacteria, that participates in the antioxidant defense system of these organisms against damage induced by organic peroxides (OHPs), such as fatty acid peroxides and peroxynitrite [1][2]. This class of peroxides is toxic due to its ability to generate free radicals, highly reactive molecules containing an unpaired electron in an atomic orbital that can cause damage to important molecules such as DNA and proteins. Because of that, these substances are frequently used by the host defense against bacteria pathogens [3][4].
The bacterial defense system is composed of enzymes like Ohr, who are capable of detoxifying and neutralizing OHPs via a redox-active disulfide bond, transforming these peroxides into unreactive alcohols [5]. Therefore, this pathway against OHPs is crucial for their survival when faced with the host defense pathway during the infection.
Ohr is unique when compared to other thiol based peroxidases, because it is very efficient in reducing OHPs but not so much when it comes to H2O2, while other peroxidases are broadspectrum, reducing OHP, H2O2 and other substrates such as fatty acid hydroperoxides and peroxynitrite with similar efficiencies. Therefore, Ohrs are specialized in OHPs, characteristic associated with the presence of a hydrophobic collar (HC) in their structure surrounding the active site, that interact more with hydrophobic substrates [2].
Ohr was described as a second system involved in bacterial resistance to OHPs by the discovery of the ohr gene from Xanthomonas campestris pv. phaseoli [2]. Furthermore, the Δohr mutant of X. campestris pv. phaseoli displayed a unique phenotype: increased and specific sensitivity to artificial OHPs but not to either H2O2 or superoxide generators. Ohr homologues from other bacteria such as Pseudomonas aeruginosa and Bacillus subtilis were also described as displaying a similar expression profile and role on OHP resistance [2].
This enzime is part of the Ohr/OsmC protein family. This group is characterized for having a Cys-based thiol-dependent peroxidase activity. Besides not having much sequence similarities, their protein structure is highly conserved, presenting two Cys residues (Cp and Cr) in their active site. The other catalytic residues (Rc and Ec) are conserved among all of the Ohr/OsmC protein family members, but were not found in Ohr-like enzymes [1][2][6].
In several bacterial genera with different lifestyles (pathogenic or non-pathogenic) other Ohr homologs and members of the Ohr/OsmC family have been found [2]. Recent studies revealed that they are not restricted to prokaryotes, being also present in eukaryotes, acquired possibly through lateral gene transfer. They found Ohr homologs in 217 species amongst non-vascular plants, animal pathogens and with especially high abundance in fungi (found in 186 species, mainly Ascomycota and Basidiomycota). However, they are absent in vertebrates and vascular plants. In most of the eukaryotic Ohr/OsmC proteins, there is a N-terminal signal sequence that predicts the cellular localization of the protein to organellar compartments such as mitochondria, peroxisome or chloroplasts. Because of that, it is proposed that the peroxidase activity of Ohr in eukaryotes is related to detoxification of endogenous sources of hydroperoxides, a different role from prokaryotes, where they are related to defense towards reactive oxygen species [6].
Regulation
OhrR, belonging to the MarR superfamily, is a Cys-based transcription factor that suppresses the expression of Ohr (Chen, 2021). OhrR can sense OHPs and, using redox-dependent structural switches, promotes a transcriptional response by binding to specific elements in the promoter of ohr gene. However, this regulation is not a universal rule, and can change depending on the microorganism. Because of its role in the virulence of some microorganisms and absence in vertebrates and vascular plants, Ohr and OhrR are interesting targets for therapeutic approaches aiming at the bacterial defense system [2].
Function
Among different bacterial species, the Ohr system can play different roles, being most of them related to OHP stress response. Despite that, it can also act in expression of virulence factors and toxic related genes; OHP detoxification and adaptive response to lipid hydroperoxide [2]. It also is known that Ohr proteins have a high affinity for oxidants such as organic hydroperoxides and peroxynitrite (Alegria et al., 2017), an aspect that is related to their structure [2].
Structure
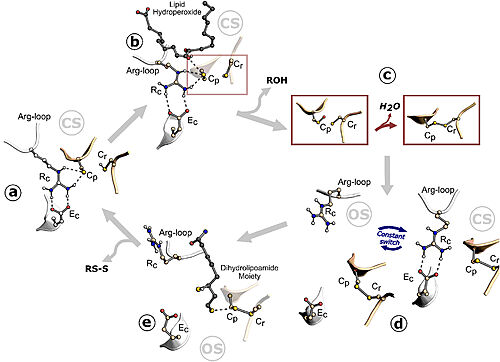
Figure 1. Ohr catalytic cycle. Image taken from Meireles et al. 2022
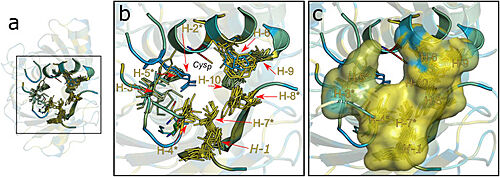
Figure 2. Hydrophobic collar (HC) surrounding the active site of Ohrs. Image taken from Meireles et al. 2022
The Ohr protein possesses an alpha/beta fold structure not found in peroxiredoxins or glutathione peroxidases (Cussiol, 2010). Ohr homologs and OsmC have motifs composed of well-conserved cysteine residues. One of these residues is part of a VCP motif, also found in peroxiredoxins, showing that Ohr can also participate in the peroxide decomposition process (Cussiol, 2003).
Ohrs have a barrel-like structure composed of folded homodimers, in which two six-stranded β-sheets wrap around two central α-helices. In this conformation, there are two active sites at opposite sides of the dimer, being two Cp residues placed in the middle of one of the central α-helices [2]. The Ohr proteins catalytic mechanism of hydroperoxide reduction is setted on a pair of redox-active cysteines, named peroxidatic (Cp) and resolving (Cr) cysteines, resembling that of the atypical 2-Cys Prxs. Ohr acts like a dimer, and the cysteine residues are placed in opposing sides of each monomer as part of two, identical active sites [6].
This structural analysis and amino acid sequence alignments showed the placement of a hydrophobic collar (HC) surrounding the active site of the protein. This part of the structure makes sense considering the fact that hydrophobicity is a common property among most Ohr substrates, such as dihydrolipoamide and lipoyl groups, both fatty acid hydroperoxides that are hydrophobic molecules [2].
The Cys (Cp) residue from Ohr forms a catalytic triad with arginine (Rc) and glutamate (Ec) within the active site of this enzyme, characterizing its function. Argc is present in a flexible loop called the Arg-loop, which moves closer to or further away from Cp. When approaching, the positive charge of the arginine induces the cysteine to adopt the deprotonated form, which is more reactive with hydroperoxides (tert-butyl hydroperoxide, for example), in the so-called "closed state". Thus, hydroperoxides enter the active site and interact with Cp, reducing them. This interaction allows cysteine to be oxidized to sulfenic acid, an unstable intermediate state that condenses with the resolving cysteine (Cr), forming disulfide bonds between the cysteines, Cp and Cr. The formation of the intramolecular disulfide bond facilitates the opening movement of the loop containing Argc, leading Ohr to adopt a conformation called the "open state" with a high affinity for its reducers. The intramolecular disulfide is then reduced, and Ohr returns to the "closed state" (Domingos et al., 2020).
Thus, in a simplified manner, due to the interactions between the amino acids of the active site, Ohr reduces its substrates to their corresponding products. To reduce more hydroperoxide molecules, it is necessary for the disulfide of this enzyme to be continuously reduced by other compounds. There is evidence that this role is performed by lipoylated proteins (Cussiol et al., 2010; Domingos et al., 2020).
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.


