User:Gustavo Sartorelli de Carvalho Rego/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
The polymerization of PHA monomers is performed by the PHA sintase/polymerase (phaC) enzyme. At first, two acetyl-CoA molecules are condensed into one acetoacetyl-CoA by a β-ketoacyl-CoA thiolase, and then reduced to (R)-3-hydroxybutyryl-CoA by the acetoacetyl-CoA dehydrogenase. Finally, phaC polymerizes the (R)-3-hydroxybutyryl-CoA molecules into the PHA polymer.<ref>Reddy, C.S.K, et al. “Polyhydroxyalkanoates: An Overview.” Bioresource Technology, vol. 87, no. 2, Apr. 2003, pp. 137–146, www.sciencedirect.com/science/article/pii/S0960852402002122, https://doi.org/10.1016/s0960-8524(02)00212-2.</ref> | The polymerization of PHA monomers is performed by the PHA sintase/polymerase (phaC) enzyme. At first, two acetyl-CoA molecules are condensed into one acetoacetyl-CoA by a β-ketoacyl-CoA thiolase, and then reduced to (R)-3-hydroxybutyryl-CoA by the acetoacetyl-CoA dehydrogenase. Finally, phaC polymerizes the (R)-3-hydroxybutyryl-CoA molecules into the PHA polymer.<ref>Reddy, C.S.K, et al. “Polyhydroxyalkanoates: An Overview.” Bioresource Technology, vol. 87, no. 2, Apr. 2003, pp. 137–146, www.sciencedirect.com/science/article/pii/S0960852402002122, https://doi.org/10.1016/s0960-8524(02)00212-2.</ref> | ||
| - | Currently, there are four known classes of phaC, that are distinguished by their primary structure, substrate specificity and subunits composition | + | Currently, there are four known classes of phaC, that are distinguished by their primary structure, substrate specificity and subunits composition <ref name='Rhem'> REHM, Bernd H. A. “Polyester Synthases: Natural Catalysts for Plastics.” Biochemical Journal, vol. 376, no. 1, 15 Nov. 2003, pp. 15–33, https://doi.org/10.1042/bj20031254.</ref><ref name='Neoh' />. Despite vast diversity, only the catalytic domain of the PhaC<sub>cn</sub>-CAT from ''Ralstonia eutropha'' H16 and the USM2 PhaC<sub>cs</sub>-CAT from the ''Chromobacterium'' sp., both being class 1 phaCs.<ref name="Neoh">Zher Neoh, Soon, et al. “Polyhydroxyalkanoate Synthase (PhaC): The Key Enzyme for Biopolyester Synthesis.” Current Research in Biotechnology, vol. 4, 2022, pp. 87–101, https://doi.org/10.1016/j.crbiot.2022.01.002.</ref> |
== Class 1 == | == Class 1 == | ||
=== Overview === | === Overview === | ||
| - | The class 1 phaCs synthetize preferentially short chain PHAs, with 3 to 5 carbon monomers, and are composed by a single enzymatic unit, with molecular mass ranging from 63 to 73 kDA.<ref>Chek, Min Fey, et al. “PHA Synthase (PhaC): Interpreting the Functions of Bioplastic-Producing Enzyme from a Structural Perspective.” Applied Microbiology and Biotechnology, vol. 103, no. 3, 3 Dec. 2018, pp. 1131–1141, https://doi.org/10.1007/s00253-018-9538-8. Accessed 14 May 2023.</ref> | + | The class 1 phaCs synthetize preferentially short chain PHAs, with 3 to 5 carbon monomers, and are composed by a single enzymatic unit, with molecular mass ranging from 63 to 73 kDA.<ref name='Neoh' /><ref name='Chek'>Chek, Min Fey, et al. “PHA Synthase (PhaC): Interpreting the Functions of Bioplastic-Producing Enzyme from a Structural Perspective.” Applied Microbiology and Biotechnology, vol. 103, no. 3, 3 Dec. 2018, pp. 1131–1141, https://doi.org/10.1007/s00253-018-9538-8. Accessed 14 May 2023.</ref> |
| - | Thanks to the X-ray cristallography data from PhaC<sub>cn</sub>-CAT and PhaC<sub>cs</sub>-CAT, it was possible to categorize the class 1 phaCs in based on their molecular organization, made of two domains: The N-terminal domain and the <scene name='10/1050300/Phac_i_catalytic_c_domain/1'>catalytic C-terminal domain</scene>. The C-terminal domain carries the <scene name='10/1050300/Phac_i_catalytic_site/2'>catalytic site, formed by the aminoacid triad (Cys-Asp-His)</scene> located deep within the <scene name='10/1050300/Phac_i_polar/1'>hydrophobic cavity</scene>, that in the closed conformation is <scene name='10/1050300/Phac_i_cap_subdomain/1'>partially covered by the Cap subdomain</scene>. | + | Thanks to the X-ray cristallography data from PhaC<sub>cn</sub>-CAT and PhaC<sub>cs</sub>-CAT, it was possible to categorize the class 1 phaCs in based on their molecular organization, made of two domains: The N-terminal domain and the <scene name='10/1050300/Phac_i_catalytic_c_domain/1'>catalytic C-terminal domain</scene>. The C-terminal domain carries the <scene name='10/1050300/Phac_i_catalytic_site/2'>catalytic site, formed by the aminoacid triad (Cys-Asp-His)</scene> located deep within the <scene name='10/1050300/Phac_i_polar/1'>hydrophobic cavity</scene>, that in the closed conformation is <scene name='10/1050300/Phac_i_cap_subdomain/1'>partially covered by the Cap subdomain</scene>.<ref name='Neoh' /><ref name='Chek' /> |
=== N-terminal domain === | === N-terminal domain === | ||
| - | The N-terminal domain has no defined function, and attempts to perform X-ray cristallography of this region have not been sucessful. Many studies have gathered evidence of possible roles that the N-terminal domain performs, such as: enzymatic activity efficiency, binding to PHA granules, substrate specificity, phaC expression, interaction with other PHA-related proteins and dimers formation and estabilization. Still, elucidation of its exact catalytic mechanism remains necessary. | + | The N-terminal domain has no defined function, and attempts to perform X-ray cristallography of this region have not been sucessful. Many studies have gathered evidence of possible roles that the N-terminal domain performs, such as: enzymatic activity efficiency, binding to PHA granules, substrate specificity, phaC expression, interaction with other PHA-related proteins and dimers formation and estabilization. Still, elucidation of its exact catalytic mechanism remains necessary.<ref name='Neoh' /> |
=== C-terminal domain === | === C-terminal domain === | ||
| Line 22: | Line 22: | ||
Regarding the <scene name='10/1050300/Phac_i_catalytic_cap_sub/1'>Cap subdomain</scene>, the LID region is extremely dynamic and flexible, having an open or closed conformation based on structural changes. Because of this, the Cap subdomain, specially the LID region, is not as conserverd in the phaCs as the α/β-hydrolase subdomain. The Cap subdomain is located after the β7 sheet, and connects with the β8 sheet from the α/β-hydrolase core. In PhaC<sub>cn</sub>, the Cap subdomain is formed by three α-helixes (α4, α5 and α6) and two β-sheets (β8 and β9). Meanwhile, PhaC<sub>cs</sub> has six α-helixes (αA, αB, αC, αD, ηA and ηB'). | Regarding the <scene name='10/1050300/Phac_i_catalytic_cap_sub/1'>Cap subdomain</scene>, the LID region is extremely dynamic and flexible, having an open or closed conformation based on structural changes. Because of this, the Cap subdomain, specially the LID region, is not as conserverd in the phaCs as the α/β-hydrolase subdomain. The Cap subdomain is located after the β7 sheet, and connects with the β8 sheet from the α/β-hydrolase core. In PhaC<sub>cn</sub>, the Cap subdomain is formed by three α-helixes (α4, α5 and α6) and two β-sheets (β8 and β9). Meanwhile, PhaC<sub>cs</sub> has six α-helixes (αA, αB, αC, αD, ηA and ηB'). | ||
| - | The cristallography of PhaC<sub>cs</sub>-CAT bound to its substrate revealed the complex assymetric dimer structure of this enzyme. The phaC dimer form can be induced by the presence the substrate. Due to the dynamic and flexible properties, specially of the LID region, the Cap subdomain is paramount in the phaC dimer formation and regulation of substrate entry and product release, since it determines the protomer's movements, regulating the change between the closed form - homodimer- and the open form - heterodimer. | + | The cristallography of PhaC<sub>cs</sub>-CAT bound to its substrate revealed the complex assymetric dimer structure of this enzyme. The phaC dimer form can be induced by the presence the substrate. Due to the dynamic and flexible properties, specially of the LID region, the Cap subdomain is paramount in the phaC dimer formation and regulation of substrate entry and product release, since it determines the protomer's movements, regulating the change between the closed form - homodimer- and the open form - heterodimer. <ref name='Neoh' /> |
=== Secondary structure === | === Secondary structure === | ||
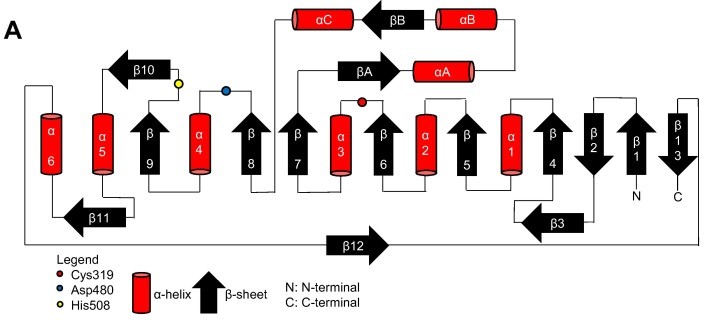
| - | Topology diagram for the catalytic domain of the PhaC<sub>cn</sub>-CAT monomer. | + | Topology diagram for the catalytic domain of the PhaC<sub>cn</sub>-CAT monomer. Image obtained from Neoh et al., 2022.<ref name='Neoh' /> |
[[Image:Proteins-cn.jpg]] | [[Image:Proteins-cn.jpg]] | ||
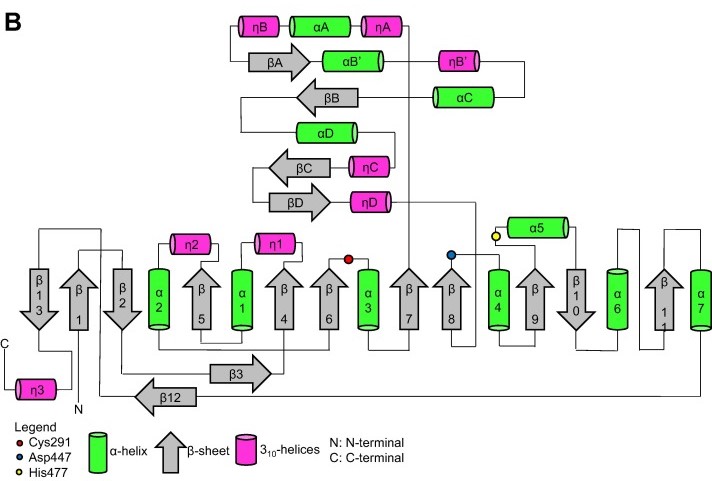
| - | Topology diagram for the catalytic domain of the PhaC<sub>cs</sub>-CAT monomer. | + | Topology diagram for the catalytic domain of the PhaC<sub>cs</sub>-CAT monomer. Image obtained from Neoh et al., 2022.<ref name='Neoh' /> |
[[Image:Proteins-cs.jpg]] | [[Image:Proteins-cs.jpg]] | ||
=== Quaternary structure === | === Quaternary structure === | ||
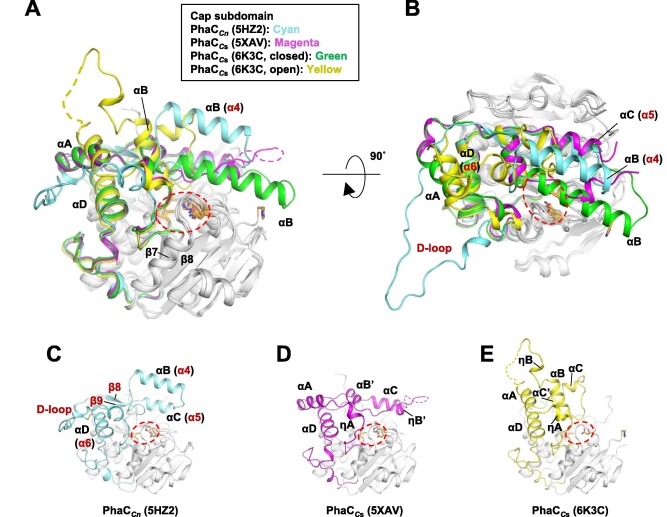
| - | Quaternary structure of PhaC<sub>cn</sub>-CAT and PhaC<sub>cs</sub>-CAT, with the Cap subdomain highlighted. | + | Quaternary structure of PhaC<sub>cn</sub>-CAT and PhaC<sub>cs</sub>-CAT, with the Cap subdomain highlighted. Image obtained from Neoh et al., 2022.<ref name='Neoh' /> |
[[Image:Proteins-quaternary.jpg]] | [[Image:Proteins-quaternary.jpg]] | ||
== Class 2 == | == Class 2 == | ||
| - | Among the 4 classes of phaC, the <scene name='10/1050300/Phac_class_ii/1'>class 2 enzymes</scene> are the only that synthetize preferentially medium chain PHA, with 6 to 14 carbon monomers. Just like the class 1, they are also formed by a single enzymatic subunit, however, its molecular mass is, in general, higher than class 1 phaC, with approximately 62 kDa. It is suggested that the class 2 molecules also have two domains. | + | Among the 4 classes of phaC, the <scene name='10/1050300/Phac_class_ii/1'>class 2 enzymes</scene> are the only that synthetize preferentially medium chain PHA, with 6 to 14 carbon monomers. Just like the class 1, they are also formed by a single enzymatic subunit, however, its molecular mass is, in general, higher than class 1 phaC, with approximately 62 kDa. It is suggested that the class 2 molecules also have two domains. <ref name='Chek' /> |
== Class 3 == | == Class 3 == | ||
Revision as of 14:30, 2 June 2024
Introduction
| |||||||||||