User:Melissa Siolin Martins/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
This structural analysis and amino acid sequence alignments showed the placement of a hydrophobic collar (HC) surrounding the active site of the protein. This part of the structure makes sense considering the fact that hydrophobicity is a common property among most Ohr substrates, such as dihydrolipoamide and lipoyl groups, both fatty acid hydroperoxides that are hydrophobic molecules. <ref name= "Meireles"/> | This structural analysis and amino acid sequence alignments showed the placement of a hydrophobic collar (HC) surrounding the active site of the protein. This part of the structure makes sense considering the fact that hydrophobicity is a common property among most Ohr substrates, such as dihydrolipoamide and lipoyl groups, both fatty acid hydroperoxides that are hydrophobic molecules. <ref name= "Meireles"/> | ||
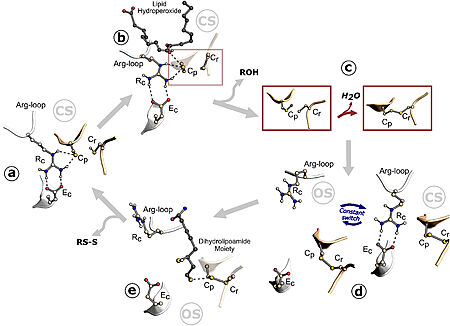
| - | The Cys (Cp) residue from Ohr forms a catalytic triad with arginine (Rc) and glutamate (Ec) within the active site of this enzyme, characterizing its function. Argc is present in a flexible loop called the Arg-loop, which moves closer to or further away from Cp. When approaching, the positive charge of the arginine induces the cysteine to adopt the deprotonated form, which is more reactive with hydroperoxides (tert-butyl hydroperoxide, for example), in the so-called "closed state". Thus, hydroperoxides enter the active site and interact with Cp, reducing them. This interaction allows cysteine to be oxidized to sulfenic acid, an unstable intermediate state that condenses with the resolving cysteine (Cr), forming disulfide bonds between the cysteines, Cp and Cr. The formation of the intramolecular disulfide bond facilitates the opening movement of the loop containing Argc, leading Ohr to adopt a conformation called the "open state" with a high affinity for its reducers. The intramolecular disulfide is then reduced, and Ohr returns to the "closed state". <ref name="Substrate">Renato M. Domingos, Raphael D. Teixeira, Ari Zeida, William A. Agudelo, Thiago G. P. Alegria, José F. da Silva Neto, Plínio S. Vieira, Mario T. Murakami, Chuck S. Farah, Dario A. Estrin, and Luis E. S. Netto | + | The Cys (Cp) residue from Ohr forms a catalytic triad with arginine (Rc) and glutamate (Ec) within the active site of this enzyme, characterizing its function. Argc is present in a flexible loop called the <scene name='10/1049474/Arg-loop/2'>Arg-loop</scene>, which moves closer to or further away from Cp. When approaching, the positive charge of the arginine induces the cysteine to adopt the deprotonated form, which is more reactive with hydroperoxides (tert-butyl hydroperoxide, for example), in the so-called "closed state". Thus, hydroperoxides enter the active site and interact with Cp, reducing them. This interaction allows cysteine to be oxidized to sulfenic acid, an unstable intermediate state that condenses with the resolving cysteine (Cr), forming disulfide bonds between the cysteines, Cp and Cr. The formation of the intramolecular disulfide bond facilitates the opening movement of the loop containing Argc, leading Ohr to adopt a conformation called the "open state" with a high affinity for its reducers. The intramolecular disulfide is then reduced, and Ohr returns to the "closed state". <ref name="Substrate">Renato M. Domingos, Raphael D. Teixeira, Ari Zeida, William A. Agudelo, Thiago G. P. Alegria, José F. da Silva Neto, Plínio S. Vieira, Mario T. Murakami, Chuck S. Farah, Dario A. Estrin, and Luis E. S. Netto |
ACS Catalysis 2020 10 (12), 6587-6602. [DOI: 10.1021/acscatal.0c01257]</ref> | ACS Catalysis 2020 10 (12), 6587-6602. [DOI: 10.1021/acscatal.0c01257]</ref> | ||
Revision as of 21:09, 2 June 2024
organic hydroperoxide resistance (Ohr)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Cussiol JR, Alegria TG, Szweda LI, Netto LE. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J Biol Chem. 2010 Jul 16;285(29):21943-50. doi: 10.1074/jbc.M110.117283. Epub, 2010 May 12. PMID:20463026 doi:http://dx.doi.org/10.1074/jbc.M110.117283
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Meireles DA, da Silva Neto JF, Domingos RM, Alegria TGP, Santos LCM, Netto LES. Ohr catalysis, phylogeny, regulation, and physiological roles. Free Radic Biol Med. 2022 May 20;185:6-24. PMID:35452809 doi:10.1016/j.freeradbiomed.2022.04.001
- ↑ Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998 May;180(10):2636-43. PMID:9573147
- ↑ Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev. 2010 Jul;4(8):118-26. PMID:22228951 doi:10.4103/0973-7847.70902
- ↑ 5.0 5.1 Chen SJ, Shu HY, Lin GH. Regulation of tert-Butyl Hydroperoxide Resistance by Chromosomal OhrR in A. baumannii ATCC 19606. Microorganisms. 2021 Mar 18;9(3):629. PMID:33803549 doi:10.3390/microorganisms9030629
- ↑ 6.0 6.1 6.2 Meireles DA, Domingos RM, Gaiarsa JW, Ragnoni EG, Bannitz-Fernandes R, da Silva Neto JF, de Souza RF, Netto LES. Functional and evolutionary characterization of Ohr proteins in eukaryotes reveals many active homologs among pathogenic fungi. Redox Biol. 2017 Aug;12:600-609. PMID:28391181 doi:10.1016/j.redox.2017.03.026
- ↑ Alegria TG, Meireles DA, Cussiol JR, Hugo M, Trujillo M, de Oliveira MA, Miyamoto S, Queiroz RF, Valadares NF, Garratt RC, Radi R, Di Mascio P, Augusto O, Netto LE. Ohr plays a central role in bacterial responses against fatty acid hydroperoxides and peroxynitrite. Proc Natl Acad Sci U S A. 2017 Jan 10;114(2):E132-E141. PMID:28028230 doi:10.1073/pnas.1619659114
- ↑ 8.0 8.1 Cussiol JR, Alves SV, de Oliveira MA, Netto LE. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J Biol Chem. 2003 Mar 28;278(13):11570-8. doi: 10.1074/jbc.M300252200. Epub 2003 , Jan 22. PMID:12540833 doi:http://dx.doi.org/10.1074/jbc.M300252200
- ↑ Renato M. Domingos, Raphael D. Teixeira, Ari Zeida, William A. Agudelo, Thiago G. P. Alegria, José F. da Silva Neto, Plínio S. Vieira, Mario T. Murakami, Chuck S. Farah, Dario A. Estrin, and Luis E. S. Netto ACS Catalysis 2020 10 (12), 6587-6602. [DOI: 10.1021/acscatal.0c01257]