Arabidospsis thaliana PrRs (pinoresinol/lariciresinol reductases)
From Proteopedia
| Line 17: | Line 17: | ||
Even so, a study conducted by Nakastsubo and collaborators (2008) showed that the recombinant AtPLRs exhibit a strong preference for pinoresinol, having almost no activity regarding lariciresinol. Because of that difference towards the family of PLRs, AtPLRs have been renamed by these authors as pinoresinol reductases (PrRs). Phylogeny analysis shows Isatis indigotica PLR1 (IiPLR1) holds the biggest relationship to AtPrR, with more than 80% of amino-acid sequence identity. However, IiPLR1 is able to use both pinoresinol and lariciresinol as substrates, meaning that the substrate specificity in AtPrR is due to few aminoacid residues alterations, causing alterations in protein structure enlightened in the next sessions (Xiao et al., 2021)<ref name="xiao" />. | Even so, a study conducted by Nakastsubo and collaborators (2008) showed that the recombinant AtPLRs exhibit a strong preference for pinoresinol, having almost no activity regarding lariciresinol. Because of that difference towards the family of PLRs, AtPLRs have been renamed by these authors as pinoresinol reductases (PrRs). Phylogeny analysis shows Isatis indigotica PLR1 (IiPLR1) holds the biggest relationship to AtPrR, with more than 80% of amino-acid sequence identity. However, IiPLR1 is able to use both pinoresinol and lariciresinol as substrates, meaning that the substrate specificity in AtPrR is due to few aminoacid residues alterations, causing alterations in protein structure enlightened in the next sessions (Xiao et al., 2021)<ref name="xiao" />. | ||
| - | |||
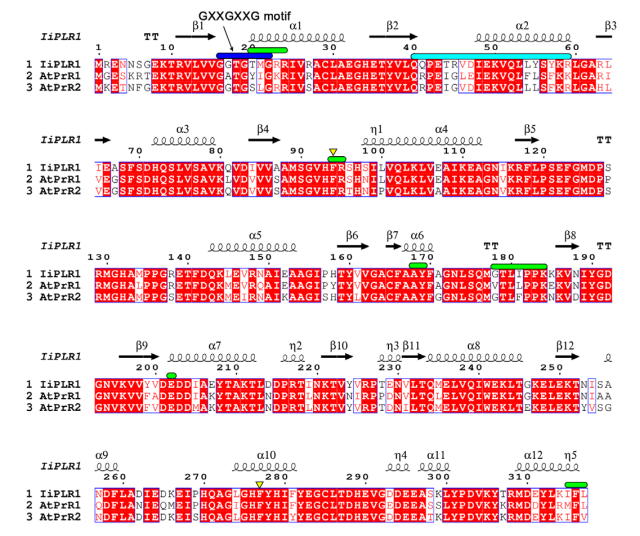
[[Image:Amino-acid sequence alignment of IiPLR and AtPrRs.png]] | [[Image:Amino-acid sequence alignment of IiPLR and AtPrRs.png]] | ||
| + | '''IMAGE 1:'''Amino-acid sequence alignment of IiPLR and AtPrRs. The secondary structure elements of IiPLR1 are placed on top. The GXXGXXG motif is indicated with blue solid lines. Regions constituting dimer interfaces of AtPrRs from Mol-A and Mol-B are indicated with green and cyan solid lines, respectively. Residues that may participate in chiral selection are indicated with yellow triangles. Species are: Ii, Isatis indigotica; At, Arabidopsis thaliana. Available in: Xiao et al., 2021<ref name="xiao" />. | ||
---- | ---- | ||
| + | |||
'''2. Protein Structure''' | '''2. Protein Structure''' | ||
| Line 37: | Line 38: | ||
In contrast with AtPrR, IiPLR β4 loops are disorder regardless of the ligand state of the protein, indicating an influence in substrate selectivity and catalysis. The β4 loop amino-acid sequence differs by one residue between IiPLR and AtPrR: at the C-terminal end of the loop, Ser98 is substituted by Asn98 in ''A. thaliana''. The interaction between Asn98 and NADPH results in a position shift of the asparagine side chain, physically limiting the β4 loop's movement in PrR. Furthermore, due to a change in the amino-acid sequence from Val46 to Leu46 in PrR1, SBD is condensed and tighter, limiting the entrance and orientation of the substrate. In PrR2, the same result is observed, and although there is no change in aminoacid residue, Val46 is pushed further into SBD, possibly owing to different dimer orientation. Thus, it is suggested that aminoacid residues at position 46 and 98 are essential for substrate selectivity (Xiao et al., 2021)<ref name="xiao" />. | In contrast with AtPrR, IiPLR β4 loops are disorder regardless of the ligand state of the protein, indicating an influence in substrate selectivity and catalysis. The β4 loop amino-acid sequence differs by one residue between IiPLR and AtPrR: at the C-terminal end of the loop, Ser98 is substituted by Asn98 in ''A. thaliana''. The interaction between Asn98 and NADPH results in a position shift of the asparagine side chain, physically limiting the β4 loop's movement in PrR. Furthermore, due to a change in the amino-acid sequence from Val46 to Leu46 in PrR1, SBD is condensed and tighter, limiting the entrance and orientation of the substrate. In PrR2, the same result is observed, and although there is no change in aminoacid residue, Val46 is pushed further into SBD, possibly owing to different dimer orientation. Thus, it is suggested that aminoacid residues at position 46 and 98 are essential for substrate selectivity (Xiao et al., 2021)<ref name="xiao" />. | ||
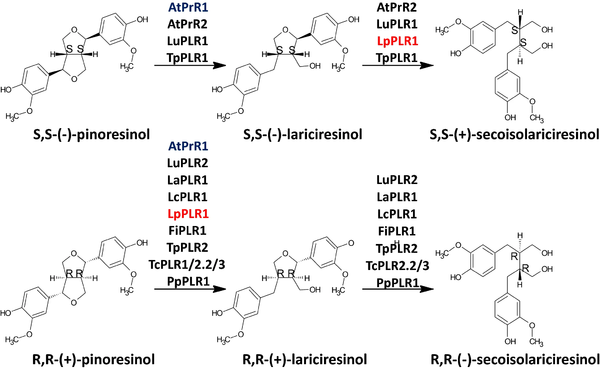
| - | [[Image:425 2019 3137 Fig2 HTML.png|600px]] | + | [[Image:425 2019 3137 Fig2 HTML.png|600px]] |
| + | '''IMAGE 2:'''Enantiospecifcity of characterized pinoresinol–lariciresinol reductases. Available in: Markulin et al., 2019<ref name="markulin" />. | ||
---- | ---- | ||
Revision as of 00:51, 3 June 2024
Página sobre a PrR1 e PrR2 de Arabidopsis feita por alunos da Biologia da USP São Paulo.
A. thaliana PrRs
| |||||||||||
4. AtPrR expression
Understanding the pattern of expression of the PLR family is key in the study of lignans, considering that the diversity of these secondary metabolites is due to the action of these enzymes, but also to their expression pattern (Xiao et al., 2021; Markulin et al., 2019)[1][2], which dictates where in the plants and when a certain lignan will be produced.
The pattern of expression of the PLRs varies from plant to plant and it can be altered by different stresses and hormones (Markulin et al., 2019). In the case of Arabidopsis thaliana, both PrR1 and PrR2 are expressed in root, but only PrR1 is also expressed in stem. Zhao and collaborators (2014)[5] have shown by co-expression analysis that PrR1 might be related to the deposition of secondary cell wall, as it clustered with genes related to: lignification, hemicellulose biosynthesis, cellulose synthesis, etc. This possibility was reinforced by transactivation assays in which the trans-activators factors MYB46 and SND1, both related to the deposition of secondary cell wall, interacted with the PrR1 promoter. On other hand, PrR2 clustered with a different set of genes, been close to genes not related to secondary cell wall deposition, as well to genes related to lignification and to the formation of the casparian strip.
What might look contradictory is the fact that PrR1 and PrR2 are coexpressed with lignin related genes and that pinoresinol, the first substrate of the PLR enzymes, is synthetized by the dimerization of two coniferyl alcohols, onde of the monomers that constitute the lignin polymer. We could imagine that the production of lignans would compete for substrates with the biosynthesis of lignin, and therefore the coexpression mentioned would be questionable. Zhao and collaborators (2014)[5].have shown that mutants of A. thaliana without the function of the PrR1 enzyme show lower content of lignin, meaning that, somehow the production of lignans is important for lignification and related to lignin polymerization.
5. References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Xiao Y, Shao K, Zhou J, Wang L, Ma X, Wu D, Yang Y, Chen J, Feng J, Qiu S, Lv Z, Zhang L, Zhang P, Chen W. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nat Commun. 2021 May 14;12(1):2828. PMID:33990581 doi:10.1038/s41467-021-23095-y
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Markulin L, Corbin C, Renouard S, Drouet S, Gutierrez L, Mateljak I, Auguin D, Hano C, Fuss E, Lainé E. Pinoresinol-lariciresinol reductases, key to the lignan synthesis in plants. Planta. 2019 Jun;249(6):1695-1714. PMID:30895445 doi:10.1007/s00425-019-03137-y
- ↑ Wang LX, Wang HL, Huang J, Chu TZ, Peng C, Zhang H, Chen HL, Xiong YA, Tan YZ. Review of lignans from 2019 to 2021: Newly reported compounds, diverse activities, structure-activity relationships and clinical applications. Phytochemistry. 2022 Oct;202:113326. PMID:35842031 doi:10.1016/j.phytochem.2022.113326
- ↑ Nakatsubo T, Mizutani M, Suzuki S, Hattori T, Umezawa T. Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J Biol Chem. 2008 Jun 6;283(23):15550-7. PMID:18347017 doi:10.1074/jbc.M801131200
- ↑ 5.0 5.1 Zhao Q, Zeng Y, Yin Y, Pu Y, Jackson LA, Engle NL, Martin MZ, Tschaplinski TJ, Ding SY, Ragauskas AJ, Dixon RA. Pinoresinol reductase 1 impacts lignin distribution during secondary cell wall biosynthesis in Arabidopsis. Phytochemistry. 2015 Apr;112:170-8. PMID:25107662 doi:10.1016/j.phytochem.2014.07.008
WARD, R. Lignans, neolignans and related compounds. v. 16, n. 1, p. 75–96, 1 jan. 1999. doi: https://doi.org/10.1039/A705992B
Proteopedia Page Contributors and Editors (what is this?)
Gabriel Fontanella Pileggi, Maisa Ganz Sanchez Sennes, Melissa Siolin Martins, Michal Harel, Angel Herraez