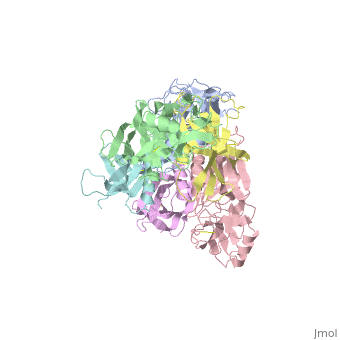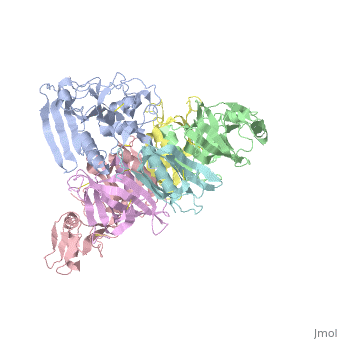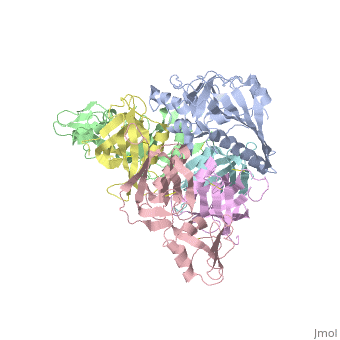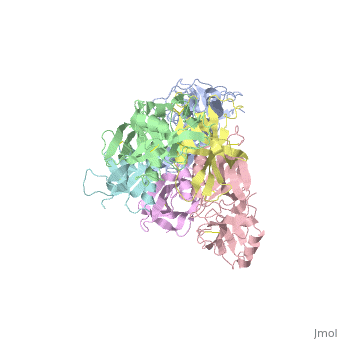We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1prt
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1prt ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1prt ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | BACKGROUND: Pertussis toxin is an exotoxin of the A-B class produced by Bordetella pertussis. The holotoxin comprises 952 residues forming six subunits (five different sequences, S1-S5). It plays an important role in the development of protective immunity to whooping cough, and is an essential component of new acellular vaccines. It is also widely used as a biochemical tool to ADP-ribosylate GTP-binding proteins in the study of signal transduction. RESULTS: The crystal structure of pertussis toxin has been determined at 2.9 A resolution. The catalytic A-subunit (S1) shares structural homology with other ADP-ribosylating bacterial toxins, although differences in the carboxy-terminal portion explain its unique activation mechanism. Despite its heterogeneous subunit composition, the structure of the cell-binding B-oligomer (S2, S3, two copies of S4, and S5) resembles the symmetrical B-pentamers of the cholera toxin and Shiga toxin families, but it interacts differently with the A-subunit. The structural similarity is all the more surprising given that there is almost no sequence homology between B-subunits of the different toxins. Two peripheral domains that are unique to the pertussis toxin B-oligomer show unexpected structural homology with a calcium-dependent eukaryotic lectin, and reveal possible receptor-binding sites. CONCLUSION: The structure provides insight into the pathogenic mechanisms of pertussis toxin and the evolution of bacterial toxins. Knowledge of the tertiary structure of the active site forms a rational basis for elimination of catalytic activity in recombinant molecules for vaccine use. | ||
| + | |||
| + | The crystal structure of pertussis toxin.,Stein PE, Boodhoo A, Armstrong GD, Cockle SA, Klein MH, Read RJ Structure. 1994 Jan 15;2(1):45-57. PMID:8075982<ref>PMID:8075982</ref> | ||
| + | |||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 1prt" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Pertussis toxin|Pertussis toxin]] | *[[Pertussis toxin|Pertussis toxin]] | ||
| + | == References == | ||
| + | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
Revision as of 05:34, 5 June 2024
THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN
| |||||||||||