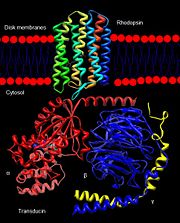
Rhodopsin
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
*'''Metarhodopsin II''' is a rhodopsin intermediate in which the retinal Schiff base is still intact but deprotonated<ref>PMID:21389988</ref>. | *'''Metarhodopsin II''' is a rhodopsin intermediate in which the retinal Schiff base is still intact but deprotonated<ref>PMID:21389988</ref>. | ||
*'''Bathorhodopsin''' and '''Lumirhodopsin''' are rhodopsin complexes with metastable intermediates of retinal<ref>PMID:21341699</ref>,<ref>PMID:11425321</ref>. | *'''Bathorhodopsin''' and '''Lumirhodopsin''' are rhodopsin complexes with metastable intermediates of retinal<ref>PMID:21341699</ref>,<ref>PMID:11425321</ref>. | ||
| - | *'''Halorhodopsin''' is a light-driven chloride pump rhodopsin that can be used to silence neural activity via light<ref>PMID:18931914</ref>. | ||
| - | *'''Archaerhodopsin''' is a light-driven proton pump<ref>PMID:30860604</ref>. | ||
| - | *'''Proteorhodopsin''' is the most abundant proton pump<ref>PMID:24060527</ref>. | ||
| - | *'''Xanthorhodopsin''' contains a cartenoid/retinal complex in which the carotenoid serves as light-harvesting antennais<ref>PMID:17571211</ref>. | ||
*'''Bestrhodopsin''' is a rhodopsin-rhodopsin-bestrophin complex<ref>PMID:35710843</ref>. | *'''Bestrhodopsin''' is a rhodopsin-rhodopsin-bestrophin complex<ref>PMID:35710843</ref>. | ||
| + | *'''Cyanorhodopsin''' is a rhodopsin found in cyanobacteria<ref>PMID:33028840</ref>. | ||
| + | *For '''Halorhodopsin''', '''Archaerhodopsin''', '''Proteorhodopsin''', '''Xanthorhodopsin''' , '''Deltarhodopsin''' see [[Bacteriorhodopsin]]. | ||
| - | See also [[Transmembrane (cell surface) receptors]] | + | |
| + | See also [[Transmembrane (cell surface) receptors]], [[Bacteriorhodopsin]] | ||
===G Protein-Coupled Receptors=== | ===G Protein-Coupled Receptors=== | ||
Current revision
| |||||||||||
References
- ↑ Hornak V, Ahuja S, Eilers M, Goncalves JA, Sheves M, Reeves PJ, Smith SO. Light activation of rhodopsin: insights from molecular dynamics simulations guided by solid-state NMR distance restraints. J Mol Biol. 2010 Feb 26;396(3):510-27. Epub 2009 Dec 11. PMID:20004206 doi:10.1016/j.jmb.2009.12.003
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002 Apr;14(2):189-95. PMID:11891118
- ↑ Tsunoda SP, Prigge M, Abe-Yoshizumi R, Inoue K, Kozaki Y, Ishizuka T, Yawo H, Yizhar O, Kandori H. Functional characterization of sodium-pumping rhodopsins with different pumping properties. PLoS One. 2017 Jul 27;12(7):e0179232. PMID:28749956 doi:10.1371/journal.pone.0179232
- ↑ Yun JH, Li X, Yue J, Park JH, Jin Z, Li C, Hu H, Shi Y, Pandey S, Carbajo S, Boutet S, Hunter MS, Liang M, Sierra RG, Lane TJ, Zhou L, Weierstall U, Zatsepin NA, Ohki M, Tame JRH, Park SY, Spence JCH, Zhang W, Schmidt M, Lee W, Liu H. Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser. Proc Natl Acad Sci U S A. 2021 Mar 30;118(13). pii: 2020486118. doi:, 10.1073/pnas.2020486118. PMID:33753488 doi:http://dx.doi.org/10.1073/pnas.2020486118
- ↑ Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011 Mar 9. PMID:21389988 doi:10.1038/nature09789
- ↑ Schapiro I, Ryazantsev MN, Frutos LM, Ferré N, Lindh R, Olivucci M. The ultrafast photoisomerizations of rhodopsin and bathorhodopsin are modulated by bond length alternation and HOOP driven electronic effects. J Am Chem Soc. 2011 Mar 16;133(10):3354-64. PMID:21341699 doi:10.1021/ja1056196
- ↑ Pan D, Mathies RA. Chromophore structure in lumirhodopsin and metarhodopsin I by time-resolved resonance Raman microchip spectroscopy. Biochemistry. 2001 Jul 3;40(26):7929-36. PMID:11425321 doi:10.1021/bi010670x
- ↑ Rozenberg A, Kaczmarczyk I, Matzov D, Vierock J, Nagata T, Sugiura M, Katayama K, Kawasaki Y, Konno M, Nagasaka Y, Aoyama M, Das I, Pahima E, Church J, Adam S, Borin VA, Chazan A, Augustin S, Wietek J, Dine J, Peleg Y, Kawanabe A, Fujiwara Y, Yizhar O, Sheves M, Schapiro I, Furutani Y, Kandori H, Inoue K, Hegemann P, Béjà O, Shalev-Benami M. Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels. Nat Struct Mol Biol. 2022 Jun;29(6):592-603. PMID:35710843 doi:10.1038/s41594-022-00783-x
- ↑ Hasegawa M, Hosaka T, Kojima K, Nishimura Y, Nakajima Y, Kimura-Someya T, Shirouzu M, Sudo Y, Yoshizawa S. A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage. Sci Rep. 2020 Oct 7;10(1):16752. PMID:33028840 doi:10.1038/s41598-020-73606-y
- ↑ 10.0 10.1 10.2 10.3 Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004 Jul;103(1):21-80. PMID:15251227 doi:10.1016/j.pharmthera.2004.05.002
- ↑ Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010 Jan;24(1):261-74. Epub 2009 Dec 17. PMID:20019124 doi:10.1210/me.2009-0473
- ↑ 12.0 12.1 12.2 Meng EC, Bourne HR. Receptor activation: what does the rhodopsin structure tell us? Trends Pharmacol Sci. 2001 Nov;22(11):587-93. PMID:11698103
- ↑ 13.0 13.1 Shieh T, Han M, Sakmar TP, Smith SO. The steric trigger in rhodopsin activation. J Mol Biol. 1997 Jun 13;269(3):373-84. PMID:9199406 doi:10.1006/jmbi.1997.1035
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 14.7 14.8 Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001 May;26(5):318-24. PMID:11343925
- ↑ 15.0 15.1 Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004 Sep 10;342(2):571-83. PMID:15327956 doi:10.1016/j.jmb.2004.07.044
- ↑ 16.0 16.1 Janz JM, Farrens DL. Assessing structural elements that influence Schiff base stability: mutants E113Q and D190N destabilize rhodopsin through different mechanisms. Vision Res. 2003 Dec;43(28):2991-3002. PMID:14611935
- ↑ 17.0 17.1 17.2 Kisselev OG. Focus on molecules: rhodopsin. Exp Eye Res. 2005 Oct;81(4):366-7. PMID:16051215 doi:10.1016/j.exer.2005.06.018
- ↑ 18.0 18.1 18.2 Verhoeven MA, Bovee-Geurts PH, de Groot HJ, Lugtenburg J, DeGrip WJ. Methyl substituents at the 11 or 12 position of retinal profoundly and differentially affect photochemistry and signalling activity of rhodopsin. J Mol Biol. 2006 Oct 13;363(1):98-113. Epub 2006 Jul 28. PMID:16962138 doi:10.1016/j.jmb.2006.07.039
- ↑ 19.0 19.1 19.2 19.3 Morris MB, Dastmalchi S, Church WB. Rhodopsin: structure, signal transduction and oligomerisation. Int J Biochem Cell Biol. 2009 Apr;41(4):721-4. Epub 2008 Aug 3. PMID:18692154 doi:10.1016/j.biocel.2008.04.025
- ↑ 20.0 20.1 20.2 20.3 20.4 Nelson, D., and Cox, M. Lehninger Principles of Biochemistry. 2008. 5th edition. W. H. Freeman and Company, New York, New York, USA. pp. 462-465.
- ↑ Hurley JB, Spencer M, Niemi GA. Rhodopsin phosphorylation and its role in photoreceptor function. Vision Res. 1998 May;38(10):1341-52. PMID:9667002
- ↑ 22.0 22.1 22.2 Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008 Jul 10;454(7201):183-7. Epub 2008 Jun 18. PMID:18563085 doi:10.1038/nature07063
- ↑ 23.0 23.1 Surya A, Knox BE. Enhancement of opsin activity by all-trans-retinal. Exp Eye Res. 1998 May;66(5):599-603. PMID:9628807 doi:10.1006/exer.1997.0453
See Also
- Bacteriorhodopsin
- G protein-coupled receptors
- Membrane proteins
- Receptor
- Transmembrane (cell surface) receptors
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Wayne Decatur, Jaime Prilusky, Joel L. Sussman, Cinting Lim