We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Riboswitch
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
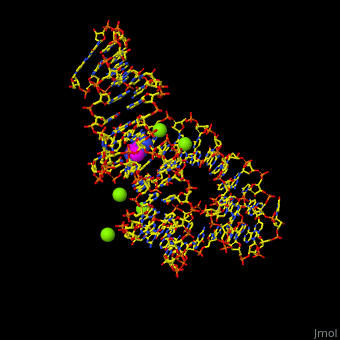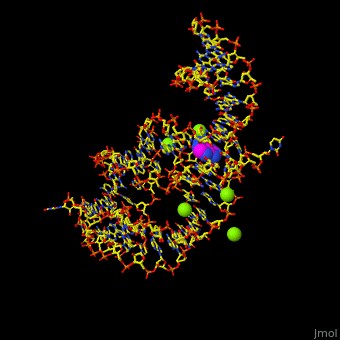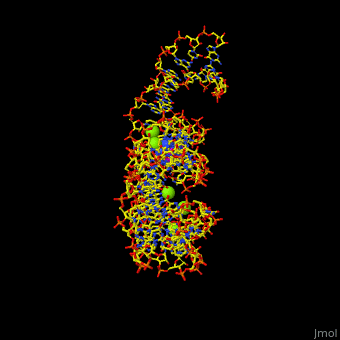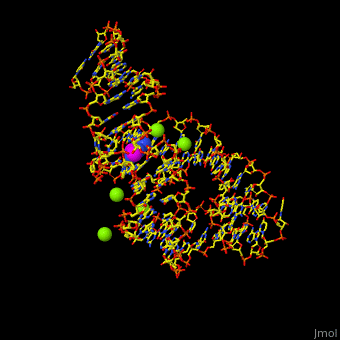
The various classes of riboswitches discovered so far are differentiated by their respective '''ligands'''. Every class of riboswitch is characterized by an aptamer (binding site) domain, which provides the site for ligand binding, and an expression platform that undergoes conformational change. The sequences and structures of aptamer domains are highly conserved, and therefore exhibit little variation among riboswitches belonging to the same class. | The various classes of riboswitches discovered so far are differentiated by their respective '''ligands'''. Every class of riboswitch is characterized by an aptamer (binding site) domain, which provides the site for ligand binding, and an expression platform that undergoes conformational change. The sequences and structures of aptamer domains are highly conserved, and therefore exhibit little variation among riboswitches belonging to the same class. | ||
| - | *M-box riboswitch couples Mg level to expression of metal transport genes<ref>PMLD:21315082</ref>. | + | *'''M-box riboswitch''' couples Mg+2 level to expression of metal transport genes<ref>PMLD:21315082</ref>. |
For details on guanine riboswitch see<br /> | For details on guanine riboswitch see<br /> | ||
* [[Guanine-Binding Riboswitch]]<br /> | * [[Guanine-Binding Riboswitch]]<br /> | ||
Revision as of 07:46, 11 August 2024
| |||||||||||
References
- ↑ Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. Science, 319(5871), 1795-1797. doi:10.1126/science.1152621
- ↑ PMLD:21315082
- ↑ Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004 Dec;11(12):1729-41. PMID:15610857 doi:S1074-5521(04)00343-6
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Karsten Theis, Joel L. Sussman