Electrostatic potential maps
From Proteopedia
(Difference between revisions)
(→Methods) |
(→PyMOL) |
||
| Line 34: | Line 34: | ||
# Menu: All, Action, remove waters. | # Menu: All, Action, remove waters. | ||
# Menu: 1pgb, Action, generate, vacuum electrostatics, protein contact potential (local). | # Menu: 1pgb, Action, generate, vacuum electrostatics, protein contact potential (local). | ||
| + | # Enter command "bg_color white". | ||
==See Also== | ==See Also== | ||
Revision as of 18:34, 25 August 2024
It is revealing to visualize the distribution of electrostatic charges, electrostatic potential, on molecular surfaces. Most protein-protein and protein-ligand interactions are largely electrostatic in nature, via hydrogen bonds and ionic interactions. Their strengths are modulated by the nature of the solvent: pure water or high ionic strength aqueous solution.
Contents |
Gallery
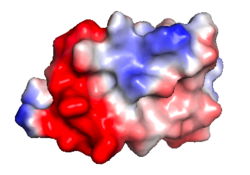
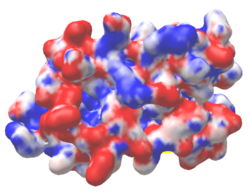
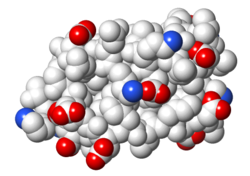
| Protein 1pgb is in the same orientation in all images. Positive + / Negative - | ||
|---|---|---|
| 
| 
|
| Electrostatic potential map rendered by PyMOL using default molecular surface probe radius 1.4 Å. Method. | Electrostatic potential map rendered by iCn3D. | Van der Waals model colored by charge with FirstGlance in Jmol. Sidechain nitrogens on Arg/Lys; oxygens on Asp/Glu. |
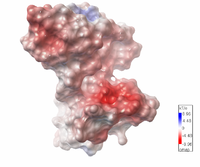
| Electrostatic potential map of 1tsj made with the Embedded Python Molecular Viewer from the Center for Computational Structural Biology of the Scripps Research Institute.
Click on the image to enlarge. |
Methods
===iCn3D
PyMOL
PyMOL has a license fee, but is free for students and educators.
- Download and install PyMOL.
- Enter command "fetch 1pgb".
- Menu: All, Action, remove waters.
- Menu: 1pgb, Action, generate, vacuum electrostatics, protein contact potential (local).
- Enter command "bg_color white".
See Also
- Electrostatic interactions in Proteopedia.
- Jmol/Electrostatic potential methods.
- Isopotential Map in Wikipedia
- Delphi Web Server
