We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1l1i
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1l1i]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Tenebrio_molitor Tenebrio molitor]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1L1I OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1L1I FirstGlance]. <br> | <table><tr><td colspan='2'>[[1l1i]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Tenebrio_molitor Tenebrio molitor]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1L1I OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1L1I FirstGlance]. <br> | ||
| - | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR</td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR, 20 models</td></tr> |
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1l1i FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1l1i OCA], [https://pdbe.org/1l1i PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1l1i RCSB], [https://www.ebi.ac.uk/pdbsum/1l1i PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1l1i ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1l1i FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1l1i OCA], [https://pdbe.org/1l1i PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1l1i RCSB], [https://www.ebi.ac.uk/pdbsum/1l1i PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1l1i ProSAT]</span></td></tr> | ||
</table> | </table> | ||
| Line 14: | Line 14: | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/l1/1l1i_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/l1/1l1i_consurf.spt"</scriptWhenChecked> | ||
| - | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/ | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> |
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1l1i ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1l1i ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
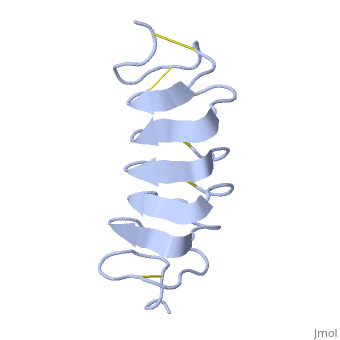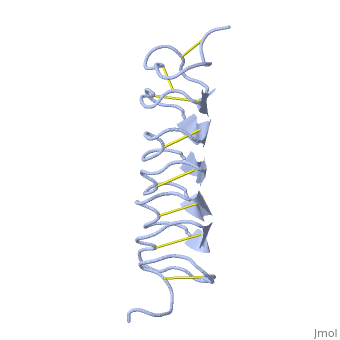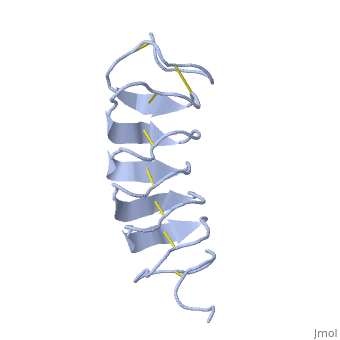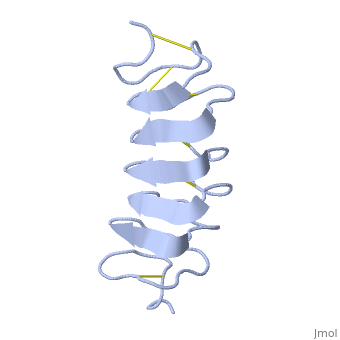
| + | Antifreeze proteins (AFPs) protect many types of organisms from damage caused by freezing. They do this by binding to the ice surface, which causes inhibition of ice crystal growth. However, the molecular mechanism of ice binding leading to growth inhibition is not well understood. In this paper, we present the solution structure and backbone NMR relaxation data of the antifreeze protein from the yellow mealworm beetle Tenebrio molitor (TmAFP) to study the dynamics in the context of structure. The full (15)N relaxation analysis was completed at two magnetic field strengths, 500 and 600 MHz, as well as at two temperatures, 30 and 5 degrees C, to measure the dynamic changes that occur in the protein backbone at different temperatures. TmAFP is a small, highly disulfide-bonded, right-handed parallel beta-helix consisting of seven tandemly repeated 12-amino acid loops. The backbone relaxation data displays a periodic pattern, which reflects both the 12-amino acid structural repeat and the highly anisotropic nature of the protein. Analysis of the (15)N relaxation parameters shows that TmAFP is a well-defined, rigid structure, and the extracted parameters show that there is similar restricted internal mobility throughout the protein backbone at both temperatures studied. We conclude that the hydrophobic, rigid binding site may reduce the entropic penalty for the binding of the protein to ice. The beta-helical fold of the protein provides this rigidity, as it does not appear to be a consequence of cooling toward a physiologically relevant temperature. | ||
| + | |||
| + | Structure and dynamics of a beta-helical antifreeze protein.,Daley ME, Spyracopoulos L, Jia Z, Davies PL, Sykes BD Biochemistry. 2002 Apr 30;41(17):5515-25. PMID:11969412<ref>PMID:11969412</ref> | ||
| + | |||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 1l1i" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
Current revision
Solution Structure of the Tenebrio molitor Antifreeze Protein
| |||||||||||