1ujw
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1ujw.gif|left|200px]] | [[Image:1ujw.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1ujw", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | | | + | or leave the SCENE parameter empty for the default display. |
| - | | | + | --> |
| - | + | {{STRUCTURE_1ujw| PDB=1ujw | SCENE= }} | |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''Structure of the complex between BtuB and Colicin E3 Receptor binding domain''' | '''Structure of the complex between BtuB and Colicin E3 Receptor binding domain''' | ||
| Line 34: | Line 31: | ||
[[Category: Zakharov, S D.]] | [[Category: Zakharov, S D.]] | ||
[[Category: Zhalnina, M V.]] | [[Category: Zhalnina, M V.]] | ||
| - | [[Category: | + | [[Category: Beta-barrel]] |
| - | [[Category: | + | [[Category: Coiled-coil]] |
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sat May 3 11:19:35 2008'' | |
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 08:19, 3 May 2008
| |||||||||
| 1ujw, resolution 2.75Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , | ||||||||
| Related: | 1nqe, 1jch | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
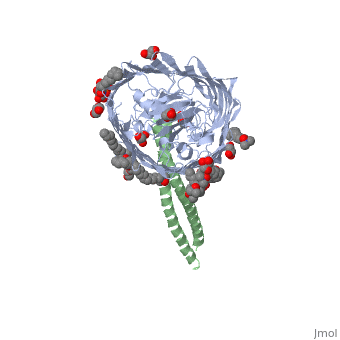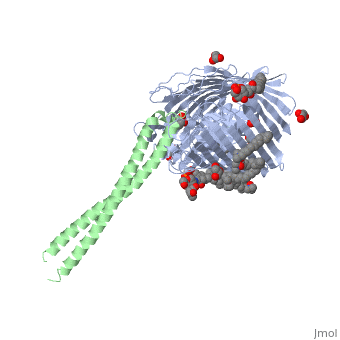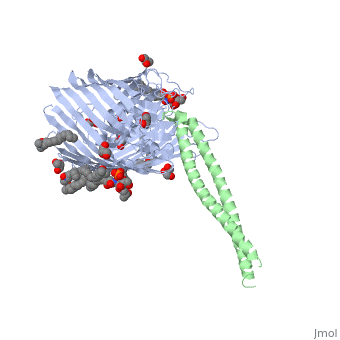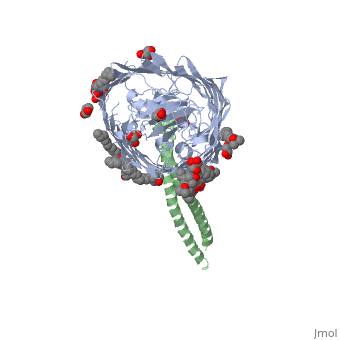
Structure of the complex between BtuB and Colicin E3 Receptor binding domain
Overview
Cellular import of colicin E3 is initiated by the Escherichia coli outer membrane cobalamin transporter, BtuB. The 135-residue 100-A coiled-coil receptor-binding domain (R135) of colicin E3 forms a 1:1 complex with BtuB whose structure at a resolution of 2.75 A is reported. Binding of R135 to the BtuB extracellular surface (DeltaG(o) = -12 kcal mol(-1)) is mediated by 27 residues of R135 near the coiled-coil apex. Formation of the R135-BtuB complex results in unfolding of R135 N- and C-terminal ends, inferred to be important for unfolding of the colicin T-domain. Small conformational changes occur in the BtuB cork and barrel domains but are insufficient to form a translocation channel. The absence of a channel and the peripheral binding of R135 imply that BtuB serves to bind the colicin, and that the coiled-coil delivers the colicin to a neighboring outer membrane protein for translocation, thus forming a colicin translocon. The translocator was concluded to be OmpF from the occlusion of OmpF channels by colicin E3.
About this Structure
1UJW is a Protein complex structure of sequences from Escherichia coli. Full crystallographic information is available from OCA.
Reference
The structure of BtuB with bound colicin E3 R-domain implies a translocon., Kurisu G, Zakharov SD, Zhalnina MV, Bano S, Eroukova VY, Rokitskaya TI, Antonenko YN, Wiener MC, Cramer WA, Nat Struct Biol. 2003 Nov;10(11):948-54. Epub 2003 Oct 5. PMID:14528295 Page seeded by OCA on Sat May 3 11:19:35 2008