|
|
| Line 1: |
Line 1: |
| | | | |
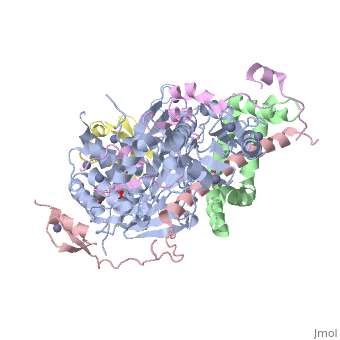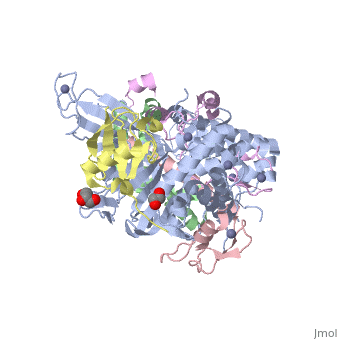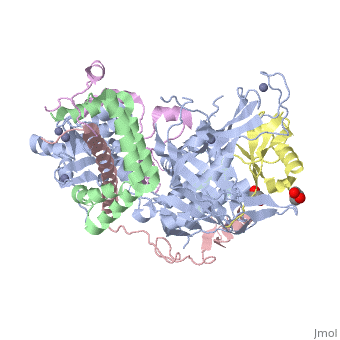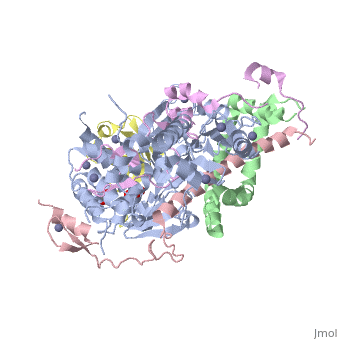
| | ==Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde== | | ==Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde== |
| - | <StructureSection load='3mhs' size='340' side='right' caption='[[3mhs]], [[Resolution|resolution]] 1.89Å' scene=''> | + | <StructureSection load='3mhs' size='340' side='right'caption='[[3mhs]], [[Resolution|resolution]] 1.89Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[3mhs]] is a 5 chain structure with sequence from [http://en.wikipedia.org/wiki/Atcc_18824 Atcc 18824] and [http://en.wikipedia.org/wiki/Human Human]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3MHS OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3MHS FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[3mhs]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3MHS OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3MHS FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.89Å</td></tr> |
| - | <tr id='NonStdRes'><td class="sblockLbl"><b>[[Non-Standard_Residue|NonStd Res:]]</b></td><td class="sblockDat"><scene name='pdbligand=GLZ:AMINO-ACETALDEHYDE'>GLZ</scene></td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=EDO:1,2-ETHANEDIOL'>EDO</scene>, <scene name='pdbligand=GLZ:AMINO-ACETALDEHYDE'>GLZ</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3mhh|3mhh]]</td></tr>
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3mhs FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3mhs OCA], [https://pdbe.org/3mhs PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3mhs RCSB], [https://www.ebi.ac.uk/pdbsum/3mhs PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3mhs ProSAT]</span></td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">UBP8, YMR223W, YM9959.05 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 ATCC 18824]), SUS1, YBR111W-A ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 ATCC 18824]), SGF11, YPL047W ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 ATCC 18824]), RPS27A, UBA52, UBB, UBC ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 HUMAN]), SGF73, YGL066W ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=4932 ATCC 18824])</td></tr>
| + | |
| - | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Ubiquitin_thiolesterase Ubiquitin thiolesterase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.1.2.15 3.1.2.15] </span></td></tr>
| + | |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3mhs FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3mhs OCA], [http://pdbe.org/3mhs PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=3mhs RCSB], [http://www.ebi.ac.uk/pdbsum/3mhs PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=3mhs ProSAT]</span></td></tr> | + | |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/UBP8_YEAST UBP8_YEAST]] Functions as histone deubiquitinating component of the transcription regulatory histone acetylation (HAT) complexes SAGA and SLIK. SAGA is involved in RNA polymerase II-dependent transcriptional regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SAGA acetylates nucleosomal histone H3 to some extent (to form H3K9ac, H3K14ac, H3K18ac and H3K23ac). SAGA interacts with DNA via upstream activating sequences (UASs). SLIK is proposed to have partly overlapping functions with SAGA. It preferentially acetylates methylated histone H3, at least after activation at the GAL1-10 locus. Together with SGF11, is required for histone H2B deubiquitination.<ref>PMID:10026213</ref> <ref>PMID:14660634</ref> <ref>PMID:15657441</ref> [[http://www.uniprot.org/uniprot/SGF11_YEAST SGF11_YEAST]] Component of the transcription regulatory histone acetylation (HAT) complex SAGA. SAGA is involved in RNA polymerase II-dependent transcriptional regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SAGA acetylates nucleosomal histone H3 to some extent (to form H3K9ac, H3K14ac, H3K18ac and H3K23ac). SAGA interacts with DNA via upstream activating sequences (UASs). SGF11 is involved in transcriptional regulation of a subset of SAGA-regulated genes. Within the SAGA complex, participates in a subcomplex with SUS1, SGF73 and UBP8 required for deubiquitination of H2B and for the maintenance of steady-state H3 methylation levels. It is required to recruit UBP8 and SUS1 into the SAGA complex.<ref>PMID:15657441</ref> <ref>PMID:15657442</ref> [[http://www.uniprot.org/uniprot/SUS1_YEAST SUS1_YEAST]] Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is involved in RNA polymerase II-dependent regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SUS1 forms a distinct functional SAGA module with UBP8, SGF11 and SGF73 required for deubiquitination of H2B and for the maintenance of steady-state H3 methylation levels. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket), by association with components of the nuclear mRNA export machinery (MEX67-MTR2 and SUB2) in the nucleoplasm and the nucleoporin NUP1 at the nuclear basket. TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery. SUS1 has also a role in mRNP biogenesis and maintenance of genome integrity through preventing RNA-mediated genome instability. Finally SUS1 has a role in response to DNA damage induced by methyl methane sulfonate (MMS) and replication arrest induced by hydroxyurea.<ref>PMID:15311284</ref> <ref>PMID:16510898</ref> <ref>PMID:16855026</ref> <ref>PMID:16760982</ref> <ref>PMID:18923079</ref> <ref>PMID:18667528</ref> <ref>PMID:18003937</ref> [[http://www.uniprot.org/uniprot/SGF73_YEAST SGF73_YEAST]] Functions as component of the transcription regulatory histone acetylation (HAT) complex SAGA. SAGA is involved in RNA polymerase II-dependent transcriptional regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SAGA acetylates nucleosomal histone H3 to some extent (to form H3K9ac, H3K14ac, H3K18ac and H3K23ac). SAGA interacts with DNA via upstream activating sequences (UASs). | + | [https://www.uniprot.org/uniprot/UBP8_YEAST UBP8_YEAST] Functions as histone deubiquitinating component of the transcription regulatory histone acetylation (HAT) complexes SAGA and SLIK. SAGA is involved in RNA polymerase II-dependent transcriptional regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SAGA acetylates nucleosomal histone H3 to some extent (to form H3K9ac, H3K14ac, H3K18ac and H3K23ac). SAGA interacts with DNA via upstream activating sequences (UASs). SLIK is proposed to have partly overlapping functions with SAGA. It preferentially acetylates methylated histone H3, at least after activation at the GAL1-10 locus. Together with SGF11, is required for histone H2B deubiquitination.<ref>PMID:10026213</ref> <ref>PMID:14660634</ref> <ref>PMID:15657441</ref> |
| | == Evolutionary Conservation == | | == Evolutionary Conservation == |
| | [[Image:Consurf_key_small.gif|200px|right]] | | [[Image:Consurf_key_small.gif|200px|right]] |
| Line 18: |
Line 15: |
| | <jmolCheckbox> | | <jmolCheckbox> |
| | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/mh/3mhs_consurf.spt"</scriptWhenChecked> | | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/mh/3mhs_consurf.spt"</scriptWhenChecked> |
| - | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> |
| | <text>to colour the structure by Evolutionary Conservation</text> | | <text>to colour the structure by Evolutionary Conservation</text> |
| | </jmolCheckbox> | | </jmolCheckbox> |
| Line 35: |
Line 32: |
| | ==See Also== | | ==See Also== |
| | *[[SAGA-associated factor|SAGA-associated factor]] | | *[[SAGA-associated factor|SAGA-associated factor]] |
| - | *[[Thioesterase|Thioesterase]] | + | *[[Thioesterase 3D structures|Thioesterase 3D structures]] |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Atcc 18824]] | + | [[Category: Homo sapiens]] |
| - | [[Category: Human]] | + | [[Category: Large Structures]] |
| - | [[Category: Ubiquitin thiolesterase]] | + | [[Category: Saccharomyces cerevisiae]] |
| - | [[Category: Berndsen, C E]] | + | [[Category: Berndsen CE]] |
| - | [[Category: Cohen, R E]] | + | [[Category: Cohen RE]] |
| - | [[Category: Datta, A B]] | + | [[Category: Datta AB]] |
| - | [[Category: Samara, N L]] | + | [[Category: Samara NL]] |
| - | [[Category: Wolberger, C]] | + | [[Category: Wolberger C]] |
| - | [[Category: Yao, T]] | + | [[Category: Yao T]] |
| - | [[Category: Zhang, X]] | + | [[Category: Zhang X]] |
| - | [[Category: Acetylation]]
| + | |
| - | [[Category: Cytoplasm]]
| + | |
| - | [[Category: Hydrolase-transcription regulator-protein binding complex]]
| + | |
| - | [[Category: Isopeptide bond]]
| + | |
| - | [[Category: Multi-protein complex]]
| + | |
| - | [[Category: Nucleus]]
| + | |
| - | [[Category: Phosphoprotein]]
| + | |
| - | [[Category: Ubl conjugation]]
| + | |
| Structural highlights
Function
UBP8_YEAST Functions as histone deubiquitinating component of the transcription regulatory histone acetylation (HAT) complexes SAGA and SLIK. SAGA is involved in RNA polymerase II-dependent transcriptional regulation of approximately 10% of yeast genes. At the promoters, SAGA is required for recruitment of the basal transcription machinery. It influences RNA polymerase II transcriptional activity through different activities such as TBP interaction (SPT3, SPT8 and SPT20) and promoter selectivity, interaction with transcription activators (GCN5, ADA2, ADA3 and TRA1), and chromatin modification through histone acetylation (GCN5) and deubiquitination (UBP8). SAGA acetylates nucleosomal histone H3 to some extent (to form H3K9ac, H3K14ac, H3K18ac and H3K23ac). SAGA interacts with DNA via upstream activating sequences (UASs). SLIK is proposed to have partly overlapping functions with SAGA. It preferentially acetylates methylated histone H3, at least after activation at the GAL1-10 locus. Together with SGF11, is required for histone H2B deubiquitination.[1] [2] [3]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
SAGA is a transcriptional coactivator complex that is conserved across eukaryotes and performs multiple functions during transcriptional activation and elongation. One role is deubiquitination of histone H2B, and this activity resides in a distinct subcomplex called the deubiquitinating module (DUBm), which contains the ubiquitin-specific protease Ubp8, bound to Sgf11, Sus1, and Sgf73. The deubiquitinating activity depends on the presence of all four DUBm proteins. We report here the 1.90 angstrom resolution crystal structure of the DUBm bound to ubiquitin aldehyde, as well as the 2.45 angstrom resolution structure of the uncomplexed DUBm. The structure reveals an arrangement of protein domains that gives rise to a highly interconnected complex, which is stabilized by eight structural zinc atoms that are critical for enzymatic activity. The structure suggests a model for how interactions with the other DUBm proteins activate Ubp8 and allows us to speculate about how the DUBm binds to monoubiquitinated histone H2B in nucleosomes.
Structural insights into the assembly and function of the SAGA deubiquitinating module.,Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C Science. 2010 May 21;328(5981):1025-9. Epub 2010 Apr 15. PMID:20395473[4]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999 Feb 26;274(9):5895-900. PMID:10026213
- ↑ Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004 Jan 16;279(3):1867-71. Epub 2003 Dec 3. PMID:14660634 doi:10.1074/jbc.C300494200
- ↑ Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005 Feb;25(3):1162-72. PMID:15657441 doi:25/3/1162
- ↑ Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010 May 21;328(5981):1025-9. Epub 2010 Apr 15. PMID:20395473 doi:10.1126/science.1190049
|