Sandbox Reserved 1849
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
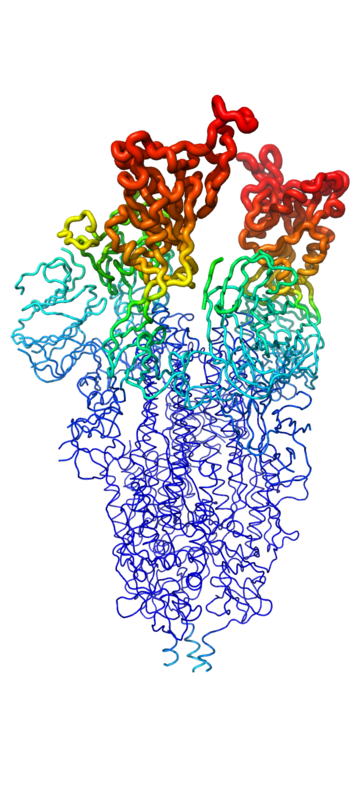
[[Image:SpikeBFactor.png|400 px|left|thumb|Figure 1. Spike protein shown in "B-Factor"; depicting mobility and flexibility of different portions. Depicted in red are the most mobile, whilst dark blue are the least mobile.]] | [[Image:SpikeBFactor.png|400 px|left|thumb|Figure 1. Spike protein shown in "B-Factor"; depicting mobility and flexibility of different portions. Depicted in red are the most mobile, whilst dark blue are the least mobile.]] | ||
| - | Throughout the entire process, the spike protein has 3 main conformations. An <scene name='10/1075251/Spike_protein_closed_spacefill/3'>inactive, "closed"</scene> conformation; an active, "open" conformation; and a <scene name='10/1075251/Spike_protein_postfusion/1'>post-fusion hairpin</scene> conformation mentioned previously<ref name="Huang">DOI:10.1038/s41401-020-0485-4 | + | Throughout the entire process, the spike protein has 3 main conformations. An <scene name='10/1075251/Spike_protein_closed_spacefill/3'>inactive, "closed"</scene> conformation; an active, "open" conformation; and a <scene name='10/1075251/Spike_protein_postfusion/1'>post-fusion hairpin</scene> conformation mentioned previously<ref name="Huang">DOI:10.1038/s41401-020-0485-4</ref><ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. In the closed conformation, the RBDs of each monomer are tucked inwards, preventing interaction. In the open conformation, however, 1 or more of these RBDs can be in the "up" conformation, meaning they are exposed and able to interact within the extracellular space. Mainly, there exits a "<scene name='10/1075251/Spike_protein_1-up_yang/1'>1-up</scene>" and "<scene name='10/1075251/Spike_protein_2-up_yang/2'>2-up</scene>" conformation in this phase<ref name="Yuan">PMID:28393837</ref><ref name="Zhang">PMID:34534731</ref>. Depicted in Figure 1, the RBDs of the spike protein have the highest mobility, which further support the many conformational changes in which they are involved. |
</p> | </p> | ||
====Function==== | ====Function==== | ||
Revision as of 18:07, 3 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
SARS-COV2 Minibinders
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020 Sep;41(9):1141-1149. doi: 10.1038/s41401-020-0485-4., Epub 2020 Aug 3. PMID:32747721 doi:http://dx.doi.org/10.1038/s41401-020-0485-4
- ↑ 4.0 4.1 4.2 Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017 Apr 10;8:15092. doi: 10.1038/ncomms15092. PMID:28393837 doi:http://dx.doi.org/10.1038/ncomms15092
- ↑ 5.0 5.1 5.2 Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021 Oct;50:173-182. PMID:34534731 doi:10.1016/j.coviro.2021.08.010