Introduction
Environmental Issues
Pollution is one of the largest environmental issues facing our globe today. It is estimated that there is 359 million tons of plastic waste, excluding all other forms of material waste, produced annually around the world. Of this plastic waste, 150-200 million tons of it ends up sitting in landfills or ends up as pollution in our natural environments. The most common form of plastic waste is poly-ethylene terephthalate, or PET, which has an annual production accumulation of six billion pounds made, generally in the form of soft drink containers and plastic water bottles. It is recycled in some aspects with around only 10% recycled. The methods that are currently being implemented are not conducive on a global scale.
Current Plastic Recycling Methodology
Strides have been taken to find a method by which PET plastics can be recycled, but many of the current employments are not conducive on a large global scale. Enzymes, specifically cutinases and hydrolases, have been a major turning point in progression of recycling. However, many of the ones studied face the issue of having a low thermal stability. PET plastics have to be broken down and/or melted at temperatures around or above 70 degrees Celsius, which the current enzymes studied cannot maintain their structure and will denature under those conditions. Some of the enzymes also do not fully break down the plastics into their original starting materials, which can be reused/recycled to make new plastic products. If the plastics cannot be fully broken down, then they cannot be recycled.
One alternative among the population of cutinases and hydrolases exists, though, that can complete the goal of recycling PET plastics at and above its transition temperature: leaf compost cutinase, or LCC. In both its wild-type and mutated versions, the LCC has become a revolutionary enzyme that could work towards the progression of ending our global environmental recycling issues.
Structure
Alpha-Beta Hydrolase Family
Leaf-branch compost bacterial cutinase features an alpha-beta hydrolase fold as its catalytic domain. The alpha-beta hydrolase fold features a chymotrypsin-like catalytic triad with a conserved histidine, a hydrophobic binding pocket, and an oxyanion hole. The primary structure contains a nucleophilic motif of G-X-Nu-X-G. The flanking glycines allow the nucleophilic region of the active site to form a tight loop called the nucleophilic elbow. Some alpha-beta hydrolase enzymes have the motif HX*4D, which allows them to exhibit acyltransferase activity. Alpha-beta hydrolase enzymes have a wide range of functions including proteolysis, signal transduction, and lipid metabolism.
Overall Topology
Leaf-branch compost bacterial cutinase, LCC, is a part of the serine hydrolase family. It is a monomer that contains a total of 258 amino acid residues, with an amphipathic structure. It has a made up of alpha helices and beta turns, which correlate to the alpha-beta hydrolase family. The active site consists of a , which is a common feature among serine hydrolases. Compared to other serine hydrolases and cutinases studied for plastic degradation, the LCC proved to be 33x more efficient.
Active Site
Structure
The active site contains a which makes aromatic pi-stacking and Van der Waals interactions with the aromatic rings in the PET ligand. Add a table with residues+monomers. There is currently no available structure of LCC with the PET ligand bound to it so the ligand position has been approximated in this model.
Table 1. list of residues in contact with PET substrate. The table shows the 15 amino acid residues in the first contact shell with the PET substrate. Residues are sorted into columns based on which monomer they are interacting with. Monomers are labeled consistently with how they were labeled in the Tournier et al. article.
| Site -2
| Site -1
| Site +1
|
| Y95
| G94
| S101
|
| F125
| T96
| F243
|
| Y127
| H164
| N246
|
| M166
| S165
|
|
| W190
| D210
|
|
| A213
| V212
|
|
|
| H242
|
|
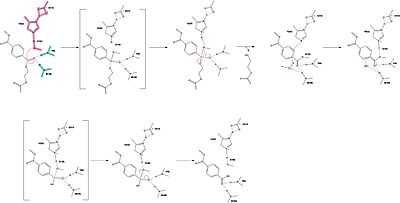
Mechanism
The mechanism involves a proton relay by the , making the catalytic S165 a good nucleophile. S165 attacks the carbonyl carbon in the -1 monomer of the PET Polymer, forming a tetrahedral intermediate (insert scene - keep old numbering?). The pi electrons move onto the carbonyl oxygen, creating an oxyanion which is stabilized by the , consisting of the backbone amide nitrogens of Y95 and M166. Then, the leaving group oxygen on the -2 monomer is protonated by H242, which is also a part of the catalytic triad (insert scene?). This facilitates the reformation of the carbonyl group upon collapse of the oxyanion and the severing of the scissile bond (insert scene).
In a second step, a water molecule is deprotonated by H242 and D210, allowing it to nucleophilically attack the carbonyl carbon, forming a tetrahedral intermediate and an oxyanion that is stabilized by the same <. H242 protonates the leaving group oxygen of S165, allowing the reformation of the carbonyl and the severing of the covalent bond to serine. The -1 monomer is released from the enzyme, and the protons are reset for further catalysis.
need higher quality image of this
Mutations
Researchers have been investigating various mutations of PET hydrolase to enhance its catalytic ability. One group of researchers, Tournier et. al., have made mutations in the PET hydrolase active site. They identified the key residues involved in the catalytic mechanism by using a model of the onto the enzyme. The site, mainly a hydrophobic pocket, contained 11 residues targeted for mutagenesis. From this, they identified that the majority of enzymes' specific activity went down; however, the mutation of the F243 to either isoleucine or tryptophan increased specific activity. Four target mutations introduced into the PET Hydrolase by Tournier et. al. demonstrated improved catalytic efficiency and thermal stability compared to its wild-type structure.
F243I/W Mutations
Mutating the active site to (in the ICCG mutant) and (in the WCCG mutant) resulted in a percent gain of specific activity of 127.5% and 118.4% respectively. More information here.
D238C/S283C Mutations
A disulfide bond was put into the structure of the LCC by mutating into cysteine disulfide scene (recopy scene link). The disulfide bond specifically replaced those residues due to the triplet acidic residue sequence E208/D238/S283 serving as a common divalent metal binding site in other cutinase enzymes. This allowed the researchers to compare the activity of the disulfide bond mutation’s activity and stability to that of the cutinases with additional metal ions that showed increased activity and stability. (Disulfide bonds are also comparable to metal cations in terms of enhancing protein structural thermal stability.) The disulfide bond proved true to enhance the thermal stability of the PET hydrolase, increasing its melting temperature to 94.5 degrees Celsius to that of the wild-type’s 84.7 degree Celsius thermal stability. A tradeoff with catalytic activity occurred, though, decreasing enzymatic activity by 28% compared to the wild type. The thermal stability seemed to be more important than activity rates if the LCC still did its intended function.
Y127G Mutation
A was introduced to residue the . The main effect of this mutation was that it showed an increase in melting temperature, but with no effect on depolymerization of the PET substrates compared to the wild type. With the addition of the glycine, the ICC/WCC mutations showed a similar or higher specific activity with a higher melting point compared to the wild type (9.3 to 13.4 degrees Celsius higher). Distances between the catalytic serine (S165) and the ligand where the scissile bond is located, as well as with the catalytic histidine (H242), decreased to enhance catalytic efficiency of LCC with new covalent interactions facilitated.
Glycosylation
were introduced in a research study completed by Abhihit N. Shirke and others with the initial intention of inducing aggregation in the leaf branch cutinase / PET hydrolase wild-type. [1] Glycosylation, as a general tool, is introduced into a protein to improve conformational stability. The specific type used in this study was N-linked side chain alteration. This means that the glycosylation sites were selected based on a starting asparagine residue followed by the sequence N-X-S or N-X-T, where X stands for any of the twenty amino acids except proline. [2] This is implemented specifically because it allows for a better ability to choose the mutation sites. The first glycosylation site followed the N-T-S pattern with . The second was with an N-A-S pattern, located nearest to the active site. The final was , exhibiting a N-D-T sequence. [1] With the combination of these glycosylation sites (without any other mutagenesis introduced to the enzyme), an overall 10-degree Celsius higher thermal stability was exhibited compared to the wild-type, with the structure of the modified PET hydrolase three times more stable. Catalytic efficiency also improved at the enzyme's known melting temperature. Even though the target point of introducing glycosylation sites was to induce aggregation in the PET hydrolase, depletion of aggregation was exhibited. The glycosylated protein took twice as long to unfold compared to the wild-type. For comparison, the threshold of 80 degrees Celsius was where the major difference in kinetic activity occurred: 85% of the glycosylated hydrolase maintained its catalytic activity, whereas the wild-type only had 50% of it working at the same temperature.[1] The first and third sites showed these trends both together and on their own as the only glycosylation sites, but the second one nearest the active site showed no change when glycosylated on its own when compared to the wild type. [1]
Biochemical Results
Improved Thermal Stability
Thermal stability is very important for enzyme-catalyzed PET degradation because the reaction must take place above the transition temperature of PET(70ºC), which allows the substrate to have optimal flexibility to fit into the active site. The disulfide bridge mutation raises the melting point of the enzyme from 84.7ºC to 94.5ºC.
Depolymerization Efficiency of Mutant LCCs
Both the ICCG and WCCG mutants designed by Tournier et al. exhibited a greater depolymerization efficiency than the wild type. The wild type LCC reached 53% depolymerization in 20 hours while both the ICCG and WCCG mutants reached 90% depolymerization in 10.5 hrs and 9.3 hrs respectively. Should this be discussed like a scientific article with these things in the conclusion or should these be discussed in the prior sections and the conclusions just talk about implications of the discovery for recycling?
Enhanced Catalytic Efficiency
The ICCG and WCCG mutations constructed by Tournier et al. showed a return to the wild type activity and beyond. The tryptophan mutation showed a 122% increase in catalysis, with an increased ability to sustain its structure at temperatures 6.2 degrees higher than the wild type (84.7 degrees Celsius). The isoleucine mutation showed a 2% decrease in activity compared to the wild type, but a thermal stability increase by 10.1 degrees Celsius.
Conclusions
Implications for Plastic Recycling
Information here