We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1849
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
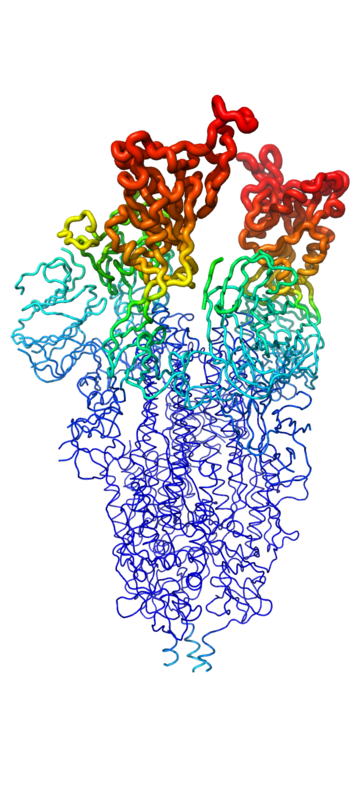
Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor, known as ACE2, and this allows the virus to remain anchored to the host for viral entry <ref name="Sang">PMID:36499120</ref>. When the spike protein binds to the receptor, ACE2 for example, the cell membrane-associated protease, protease serine 2 TMPRSS2 promotes viral entry by activating the spike protein <ref name="Huang">PMID:32747721</ref>. The activated spike protein is able to cleave itself into S1 and S2 subunits <ref name="Huang">PMID:32747721</ref>. The S2 subunit is in charge of viral entry and does this through conformational changes <ref name="Huang">PMID:32747721</ref>. The S2 subunit will insert it's FP domain into the host cell's membrane, and this will trigger an interaction with the HR2 domain and HR1 trimer to form the 6-helical bundle to bring the viral envelope and cell membrane in close enough distance for viral fusion and ultimately viral entry <ref name="Huang">PMID:32747721</ref>. Once the virus is within the host cell, it is able to translate viral proteins, eliciting an immune response and spreading the viral particles throughout the body <ref name="Huang">PMID:32747721</ref>. | Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor, known as ACE2, and this allows the virus to remain anchored to the host for viral entry <ref name="Sang">PMID:36499120</ref>. When the spike protein binds to the receptor, ACE2 for example, the cell membrane-associated protease, protease serine 2 TMPRSS2 promotes viral entry by activating the spike protein <ref name="Huang">PMID:32747721</ref>. The activated spike protein is able to cleave itself into S1 and S2 subunits <ref name="Huang">PMID:32747721</ref>. The S2 subunit is in charge of viral entry and does this through conformational changes <ref name="Huang">PMID:32747721</ref>. The S2 subunit will insert it's FP domain into the host cell's membrane, and this will trigger an interaction with the HR2 domain and HR1 trimer to form the 6-helical bundle to bring the viral envelope and cell membrane in close enough distance for viral fusion and ultimately viral entry <ref name="Huang">PMID:32747721</ref>. Once the virus is within the host cell, it is able to translate viral proteins, eliciting an immune response and spreading the viral particles throughout the body <ref name="Huang">PMID:32747721</ref>. | ||
===COVID-19 Viral Infection Interruption=== | ===COVID-19 Viral Infection Interruption=== | ||
| - | The primary goal of the mini binders is to prevent the spike proteins from binding to ACE2, and when the mini binders are bound to the spike protein, the virus is unable to anchor itself to the host protein <ref name="Longxing">PMID:32907861</ref>. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response <ref name="Longxing">PMID:32907861</ref>. Targeting this specific interaction between the COVID-19 spike protein has proven effective and is hopeful target for future therapeutics to treat the virus <ref name="Huang">PMID:32747721</ref>. LCB1 proved to be quite effective at weakening the immune response, compared to the other mini binders, which can be explained by the | + | The primary goal of the mini binders is to prevent the spike proteins from binding to ACE2, and when the mini binders are bound to the spike protein, the virus is unable to anchor itself to the host protein <ref name="Longxing">PMID:32907861</ref>. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response <ref name="Longxing">PMID:32907861</ref>. Targeting this specific interaction between the COVID-19 spike protein has proven effective and is hopeful target for future therapeutics to treat the virus <ref name="Huang">PMID:32747721</ref>. LCB1 proved to be quite effective at weakening the immune response, compared to the other mini binders, which can be explained by the binding interface between the spike protein and LCB1 <ref name="Longxing">PMID:32907861</ref>. |
Revision as of 18:57, 10 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
SARS-COV2 Minibinders
| |||||||||||
References
[3] [4] [5] [6] [8] [7] [9] [10] [11]
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020 Oct 23;370(6515):426-431. PMID:32907861 doi:10.1126/science.abd9909
- ↑ 4.0 4.1 Case JB, Chen RE, Cao L, Ying B, Winkler ES, Johnson M, Goreshnik I, Pham MN, Shrihari S, Kafai NM, Bailey AL, Xie X, Shi PY, Ravichandran R, Carter L, Stewart L, Baker D, Diamond MS. Ultrapotent miniproteins targeting the SARS-CoV-2 receptor-binding domain protect against infection and disease. Cell Host Microbe. 2021 Jul 14;29(7):1151-1161.e5. PMID:34192518 doi:10.1016/j.chom.2021.06.008
- ↑ 5.0 5.1 Sang P, Chen YQ, Liu MT, Wang YT, Yue T, Li Y, Yin YR, Yang LQ. Electrostatic Interactions Are the Primary Determinant of the Binding Affinity of SARS-CoV-2 Spike RBD to ACE2: A Computational Case Study of Omicron Variants. Int J Mol Sci. 2022 Nov 26;23(23):14796. PMID:36499120 doi:10.3390/ijms232314796
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 6.12 Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020 Sep;41(9):1141-1149. doi: 10.1038/s41401-020-0485-4., Epub 2020 Aug 3. PMID:32747721 doi:http://dx.doi.org/10.1038/s41401-020-0485-4
- ↑ 7.0 7.1 7.2 7.3 Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021 Oct;50:173-182. PMID:34534731 doi:10.1016/j.coviro.2021.08.010
- ↑ 8.0 8.1 8.2 Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017 Apr 10;8:15092. doi: 10.1038/ncomms15092. PMID:28393837 doi:http://dx.doi.org/10.1038/ncomms15092
- ↑ 9.0 9.1 9.2 Kuba K, Yamaguchi T, Penninger JM. Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19. Front Immunol. 2021 Dec 22;12:732690. PMID:35003058 doi:10.3389/fimmu.2021.732690
- ↑ 10.0 10.1 Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005 Aug;11(8):875-9. PMID:16007097 doi:10.1038/nm1267
- ↑ 11.0 11.1 Valetti F, Gilardi G. Improvement of biocatalysts for industrial and environmental purposes by saturation mutagenesis. Biomolecules. 2013 Oct 8;3(4):778-811. PMID:24970191 doi:10.3390/biom3040778