We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
===DA_20_10=== | ===DA_20_10=== | ||
====Q162R==== | ====Q162R==== | ||
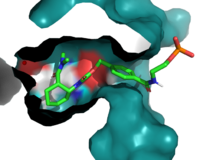
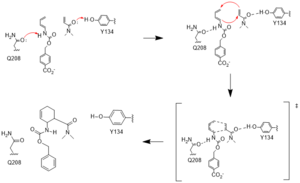
| - | Residue 162 resides near the top of the binding entrance to the enzyme, and is within 3A in most models on the enzyme. It can act as a hydrogen bond donor to the terminal phosphate on the ligand when in proximity. To increase this interaction, the group chose a Q to <scene name='10/1075254/Q_to_r/1'>R mutation</scene>, which decreased the length of the potential hydrogen bond to within 2.5A, increasing the strength of the interaction. | + | Residue 162 resides near the top of the binding entrance to the enzyme, and is within 3A in most models on the enzyme. It can act as a hydrogen bond donor to the terminal phosphate on the ligand when in proximity. To increase this interaction, the group chose a Q to <scene name='10/1075254/Q_to_r/1'>R mutation</scene>, which decreased the length of the potential hydrogen bond to within 2.5A, increasing the strength of the interaction. |
| + | ====A285N==== | ||
| + | Residue 285, as follows, is also buried within the binding pocket. The group introduced this mutation to increase steric hindrance with the catalytic tyrosine, reducing the number of rotamers the residue has to increase the reactivity of the enzyme by lowering the distance between Y134 and the ligand. | ||
===CE20=== | ===CE20=== | ||
In this generation, it was found that the most catalytically efficient models had mutated T34, P48, and R56 to <scene name='10/1075254/Ce_20_mutations/4'>I43,L48, and S56</scene>. These mutations further tightened the binding pocket and create a more hydrophobic environment. | In this generation, it was found that the most catalytically efficient models had mutated T34, P48, and R56 to <scene name='10/1075254/Ce_20_mutations/4'>I43,L48, and S56</scene>. These mutations further tightened the binding pocket and create a more hydrophobic environment. | ||
Revision as of 19:48, 10 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Diels-Alderase
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
Taylor Donahue Kate Thuma Micah Zile