User:Elizabeth Yowell/ SandboxFinal
From Proteopedia
| Line 50: | Line 50: | ||
==Binding Site and Interactions== | ==Binding Site and Interactions== | ||
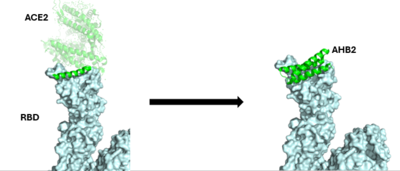
| - | <scene name='10/1075219/ | + | In order to best target the RBD of the spike protein, the minibinders reveal a wide range of interactions to compete with <scene name='10/1075219/Ace2/1'>ACE2 binding</scene>. ACE2's main interactions with the spike protein include D30, H34, Y41, D38, and E35 of ACE2 with K417, Y453, Q498, K353, and Q493 of the spike protein, respectively. |
| - | <scene name='10/1075219/ | + | The first design method, Rosetta blueprint, created AHB2 based on the single interacting helix of ACE2. With <scene name='10/1075219/Ahb2/2'>AHB2 binding</scene>, we see two alpha helices mimicking ACE2. Hydrogen bonding interactions between N36, D11, K43, E41, and E30 of the minibinder interact with residues K417, R403, Y449, Q493, and N487, respectively, in the spike protein. One interesting result of the Rosetta blueprint design was the addition of the second helix to interact with the spike protein. It is assumed that this addition further stabilizes the interaction, increasing stability of the minibinder:spike complex. |
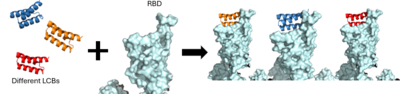
| - | <scene name='10/1075219/ | + | De novo designed proteins, as discussed previously, focused on computational design to determine residues best able to interact with the spike protein. We will focus on LCB1 and LCB3. <scene name='10/1075219/Lcb1/4'>LCB1 binding</scene> reveals hydrogen bonding between D30 of the minibinder and both K417 and R403 of the spike protein, in addition to D17 and R14 of the minibinder interacting with Q493 of the spike protein. Similarly, <scene name='10/1075219/Lcb3/5'>LCB3 binding</scene> reveals hydrogen bonding between D11 of the minibinder to K417 and R403 of the spike protein. |
| - | + | Within all four binding sites, we see two spike protein residues interacting with all four minibinders: K417 and Q493. This suggests that these resides are critical for binding. | |
==Inhibitor Differences== | ==Inhibitor Differences== | ||
Revision as of 18:01, 13 April 2025
Contents |
Engineered Protein Inhibitors for SARS-CoV-2 Entry
| |||||||||||
References
Cao, L., Goreshnik, I., Coventry, B., Case, J.B., Miller, L., Kozodoy, L., Chen, R.E., Carter, L., Walls, A.C., Park, Y., Strauch, E., Stewart, L., Diamond, M.S., Veesler, D., & Baker, D. De novo design of picomolar SARS-CoV-2 mini protein inhibitors. Science 370, 426-431 (2020). https://doi.org/10.1126/science.abd9909
https://www.who.int/europe/emergencies/situations/covid-19
https://www.nature.com/articles/s41580-021-00418-x#citeas
https://www.science.org/doi/10.1126/science.abd9909
Zhang, Haoran et al. Advances in developing ACE2 derivatives against SARS-CoV-2. The Lancet Microbe, Volume 4, Issue 5, e369 - e378 (2023). https://doi.org/10.1016/S2666-5247(23)00011-3
PDB Files
[1]https://www.rcsb.org/structure/7UHB
Student Contributors
- Giavanna Yowell
- Shea Bailey
- Matthew Pereira