We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1845
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | {{Sandbox_Reserved_CH462_Biochemistry_II_2025}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | |
| - | = | + | =Leaf Branch Compost Cutinase= |
| - | <StructureSection load=' | + | <StructureSection load='4EB0_with_substrate.pdb' size='340' side='right' caption='Original Structure of LCC' scene='10/1075246/4eb0_in_pink/2'> |
| - | + | '''Ashley Callaghan/Sandbox1''' | |
| - | + | ==Introduction== | |
| + | Leaf branch compost [https://en.wikipedia.org/wiki/Cutinase cutinase] <scene name='10/1075246/4eb0_in_pink/2'>(LCC)</scene> is a versatile enzyme that can break down both natural plant polymers and synthetic plastics.<ref name="Tournier">PMID:32269349</ref> It was discovered in a [https://en.wikipedia.org/wiki/Compost compost] heap, and it originally evolved to degrade cutin, the protective biopolymer in plant surfaces. LCC has also shown high efficiency in hydrolyzing [https://en.wikipedia.org/wiki/Polyethylene_terephthalate polyethylene terephthalate] (PET), which is a widely used plastic that contributes to global pollution. Unlike many other PET-degrading enzymes, LCC is thermostable ''and'' has a high catalytic efficiency, which means it can function at temperatures that are optimal for industrial recycling processes. By breaking PET into its monomers, LCC promotes closed-loop recycling of plastic waste and reduces environmental accumulation. | ||
| + | <ref name="Kolattukudy">PMID:17779010</ref> | ||
| + | <ref name="Burgin">PMID:38538850</ref> | ||
| - | == | + | == Function == |
| + | LCC catalyzes the hydrolysis of the ester bonds in PET polymers <scene name='10/1075247/2he_met_3/1'>2-HE(MET)3</scene> and breaks them down into their constituent monomers: terephthalic acid and ethylene glycol. The enzyme operates through a catalytic triad that consists of S165, D210, and H242. Using these residues, LCC can perform nucleophilic attacks on the carbonyl carbon atoms of the ester bonds in PET. During catalysis, the substrate binds in an elongated, predominantly hydrophobic groove present in the enzyme's structure. | ||
| + | LCC functions optimally at elevated temperatures (around 65-72°C), which approaches the [https://en.wikipedia.org/wiki/Glass_transition glass transition] temperature of PET. This temperature range maximizes PET chain mobility and makes the polymer more accessible to enzymatic action. The enzyme is remarkably thermostable compared to other PET hydrolases, with a melting temperature of 84.7°C. This property allows it to remain functional under these high-temperature conditions. Also unlike other PET hydrolases such as Is-PETase, BTA1, BTA2, and FsC, LCC has substantially higher catalytic efficiency. Specifically, LCC has an initial PET-specific depolymerization rate of 93.2 mg TAeq. h⁻¹ mg⁻¹ enzyme at 65°C with amorphous PET. This means that it us at least 33 times more efficient than other tested enzymes.<ref name="Tournier">PMID:32269349</ref> | ||
| + | LCC's function is limited by PET [https://en.wikipedia.org/wiki/Crystallization_of_polymers crystallinity], as the enzyme can more effectively hydrolyze amorphous regions of the polymer. As PET crystallinity increases during the depolymerization reaction (due to exposure to elevated temperatures), the enzyme's efficiency decreases. This limits complete depolymerization unless optimal conditions and enzyme variants are used. | ||
| - | < | + | == Relevance == |
| + | With global plastic production reaching approximately 299 million tons annually, the need for effective waste management solutions is urgent. Enzymatic degradation is an alternative to conventional recycling methods that are often inefficient and taxing on the environment. One of the primary challenges in plastic waste management is the volume of mismanaged plastic entering marine environments. In 2010 alone, an estimated 31.9 million metric tons of plastic waste were classified as mismanaged, with a substantial portion ending up in the oceans. This causes harm to marine ecosystems, physical injury to wildlife, and disruption of food chains.<ref name="Landrigan">PMID:33354517</ref> | ||
| + | Integrating LCC into existing waste management systems could substantially reduce the PET waste that enters the environment. Research suggests that a 77% reduction in mismanaged plastic waste could lower the annual input of plastic into the ocean to between 2.4 and 6.4 million metric tons by 2025. | ||
| + | LCC hydrolyzes PET into its constituent monomers, which also supports the principles of a circular economy, where materials are reused rather than discarded. Enzymatic degradation allows for the production of biologically recycled PET with properties that are comparable to virgin materials.<ref name="Jambeck">PMID:25678662</ref> | ||
== Structural Overview == | == Structural Overview == | ||
| + | <scene name='10/1075246/4eb0_helix_sheet/1'>Secondary Structures</scene> | ||
=== Catalytic Triad === | === Catalytic Triad === | ||
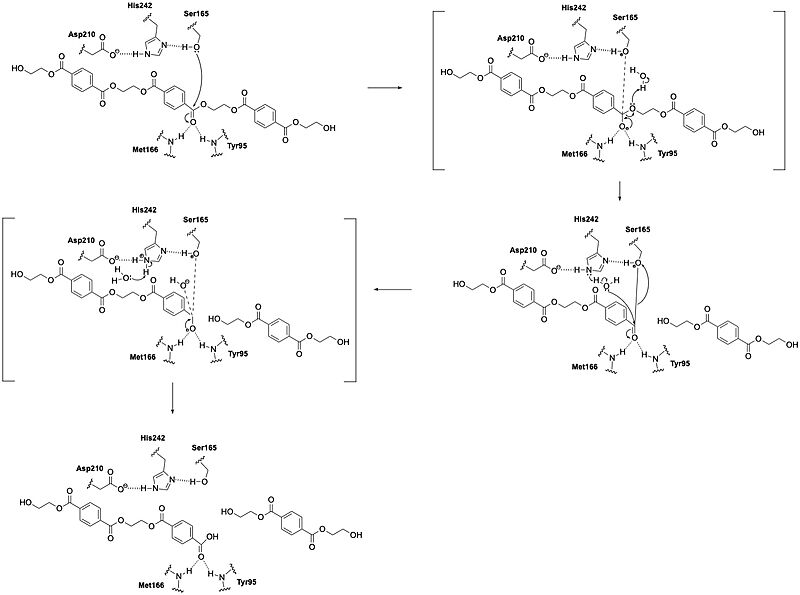
| + | LCC catalyzes the breakdown of PET using a classic serine hydrolase mechanism involving a <scene name='10/1075246/Catalytic_triad3/1'>catalytic triad</scene> of Ser165, His242, and Asp210. The reaction begins when His242 deprotonates Ser165, which activates it as a nucleophile. Ser165 then attacks the carbonyl carbon of an ester bond in the PET polymer. This forms a tetrahedral intermediate. This intermediate is stabilized by an oxyanion hole. The oxyanion hole is formed by the backbone amides of Met166 and Tyr95. The intermediate collapses; one product is released and an acyl-enzyme intermediate is formed. A water molecule, activated by His242, then attacks the acyl-enzyme, releasing the second product and resetting the enzyme’s active site. | ||
| + | [[Image:CH464 PyMOL Presentation Mechanism.jpeg|800 px|right|thumb|Figure 1: Mechanism]] | ||
| - | <scene name='10/ | + | === Ligand Binding Pocket === |
| + | The <scene name='10/1075246/4eb0_with_colored_ligand_stick/1'>substrate-binding site</scene> of LCC is a long, mainly hydrophobic groove that accommodates PET chains. This groove includes three subsites—designated −2, −1, and +1—that interact with specific PET units near the scissile ester bond. Hydrophobic residues such as F125, V212, M166, and F243 line the groove and facilitate binding by interacting with the aromatic rings of the PET molecule. These interactions help align the substrate in the correct position for catalysis. | ||
| + | [[Image:Final rayed image of binding pocket.png|400 px|right|thumb|Figure 2: Ser, His, Asp Catalytic Triad]] | ||
| + | <scene name='10/1075246/4eb0_surface_w_stick_ligand/1'>Molecular surface view of enzyme-ligand interaction</scene> | ||
| + | <scene name='10/1075246/4eb0_hydrophobicity_ligand/1'>Cartoon representation of enzyme-ligand interaction</scene> | ||
| - | == | + | == Mutation Sites of Interest == |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=== F243 === | === F243 === | ||
<scene name='10/1075247/F243/3'>F243</scene> | <scene name='10/1075247/F243/3'>F243</scene> | ||
| - | <scene name='10/1075247/I243/3'> | + | <scene name='10/1075247/I243/3'>F243I</scene> |
F243W | F243W | ||
| - | The original side chain is phenylalanine | + | The original side chain at position 243 is phenylalanine, which is located 3.6 Å from the ligand. Two mutations at this position—F243I (isoleucine) and F243W (tryptophan)—increase the catalytic activity of the enzyme. The F243I mutation replaces phenylalanine with isoleucine, a smaller side chain that allows the ligand to sit closer. This reduces the ligand distance to 3.0 Å. This tighter interaction likely improves substrate binding. The F243W mutation introduces tryptophan, which has a bulkier, nitrogen-containing aromatic side chain. Tryptophan brings the ligand slightly closer at 3.2 Å and introduces potential for new interactions, such as hydrogen bonding or π-stacking. Both mutations result in improved catalytic performance. The F243I mutation leads to a 27.5% increase in activity, while the F243W mutation results in a 17.5% increase, compared to the wild-type enzyme. |
| - | + | ||
=== T96 === | === T96 === | ||
<scene name='10/1075247/T96/3'>T96</scene> | <scene name='10/1075247/T96/3'>T96</scene> | ||
| - | + | T96M | |
| - | The | + | The mutation of threonine to methionine at position 96 (M96) increases the protein's thermostability. Threonine has a small, polar side chain, which makes it hydrophilic and less stable at higher temperatures. Methionine, however, has a larger, nonpolar, and hydrophobic side chain, which strengthens hydrophobic interactions in the protein’s core. These stronger internal interactions help stabilize the protein and help it to maintain its folded structure at higher temperatures. As a result, the melting point increases from 84.7°C for the wild-type protein to 87.4°C for the M96 mutant. |
=== Y127 === | === Y127 === | ||
<scene name='10/1075247/Y127/1'>Y127</scene> | <scene name='10/1075247/Y127/1'>Y127</scene> | ||
| - | <scene name='10/1075247/G127/1'> | + | <scene name='10/1075247/G127/1'>Y127G</scene> |
| - | The | + | The mutation of tyrosine to glycine at position 127 (Y127G) also increases the protein's thermostability. The mutant melting point is increased to 87.0°C. Tyrosine has a bulky, rigid aromatic side chain that can cause structural strain. Glycine is the smallest amino acid and lacks a side chain, so it provides greater flexibility to the protein. This mutation reduces steric hindrance and relieves strain in the protein structure, therefore allowing it to be more adaptable and stable at higher temperatures. By increasing flexibility, the Y127G mutation helps the protein maintain its folded structure under heat stress. |
=== N246 === | === N246 === | ||
<scene name='10/1075247/N246/1'>N246</scene> | <scene name='10/1075247/N246/1'>N246</scene> | ||
| - | + | N246D | |
| - | + | N246M | |
| - | The | + | The asparagine side chain is mutated to aspartic acid and methionine to increase thermostability. The wild-type protein has a melting point of 84.7°C. The N246D mutation (asparagine to aspartic acid) replaces a polar neutral side chain with a negatively charged one, which potentially increases electrostatic interactions or salt bridges that stabilize the protein. This results in a melting point of 87.9°C. The N246M mutation (asparagine to methionine) introduces a bulkier, hydrophobic side chain. This increases the internal packing of the protein core. This mutant has a melting point of 88.0°C. |
=== S283 & D238 === | === S283 & D238 === | ||
| - | <scene name='10/ | + | <scene name='10/1075248/S283-d238/1'>S283 and D238</scene> |
| - | <scene name='10/ | + | <scene name='10/1075248/C283-c238/1'>S283C and D238C</scene> |
| - | + | Two wild-type residues, S283 and D238, were engineered to form a disulfide bond by replacing them with cysteine. The wild-type protein has a melting point of 84.7°C, while the cysteine mutation increased the melting point to 94.5°C, a 9.8°C improvement—higher than any other mutations. However, this increase in stability was accompanied by a 28% decrease in enzymatic activity compared to the wild-type. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | < | + | <references/> |
| + | |||
| + | ==Student Contributors== | ||
| + | Ashley Callaghan | ||
| + | Rebecca Hoff | ||
| + | Simone McCowan | ||
Revision as of 02:35, 14 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Leaf Branch Compost Cutinase
| |||||||||||
References
- ↑ 1.0 1.1 Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Kolattukudy PE. Biopolyester membranes of plants: cutin and suberin. Science. 1980 May 30;208(4447):990-1000. PMID:17779010 doi:10.1126/science.208.4447.990
- ↑ Burgin T, Pollard BC, Knott BC, Mayes HB, Crowley MF, McGeehan JE, Beckham GT, Woodcock HL. The reaction mechanism of the Ideonella sakaiensis PETase enzyme. Commun Chem. 2024 Mar 27;7(1):65. PMID:38538850 doi:10.1038/s42004-024-01154-x
- ↑ Landrigan PJ, Stegeman JJ, Fleming LE, Allemand D, Anderson DM, Backer LC, Brucker-Davis F, Chevalier N, Corra L, Czerucka D, Bottein MD, Demeneix B, Depledge M, Deheyn DD, Dorman CJ, Fénichel P, Fisher S, Gaill F, Galgani F, Gaze WH, Giuliano L, Grandjean P, Hahn ME, Hamdoun A, Hess P, Judson B, Laborde A, McGlade J, Mu J, Mustapha A, Neira M, Noble RT, Pedrotti ML, Reddy C, Rocklöv J, Scharler UM, Shanmugam H, Taghian G, van de Water JAJM, Vezzulli L, Weihe P, Zeka A, Raps H, Rampal P. Human Health and Ocean Pollution. Ann Glob Health. 2020 Dec 3;86(1):151. PMID:33354517 doi:10.5334/aogh.2831
- ↑ Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL. Marine pollution. Plastic waste inputs from land into the ocean. Science. 2015 Feb 13;347(6223):768-71. PMID:25678662 doi:10.1126/science.1260352
Student Contributors
Ashley Callaghan Rebecca Hoff Simone McCowan