We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1846
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
=== Catalytic Triad === | === Catalytic Triad === | ||
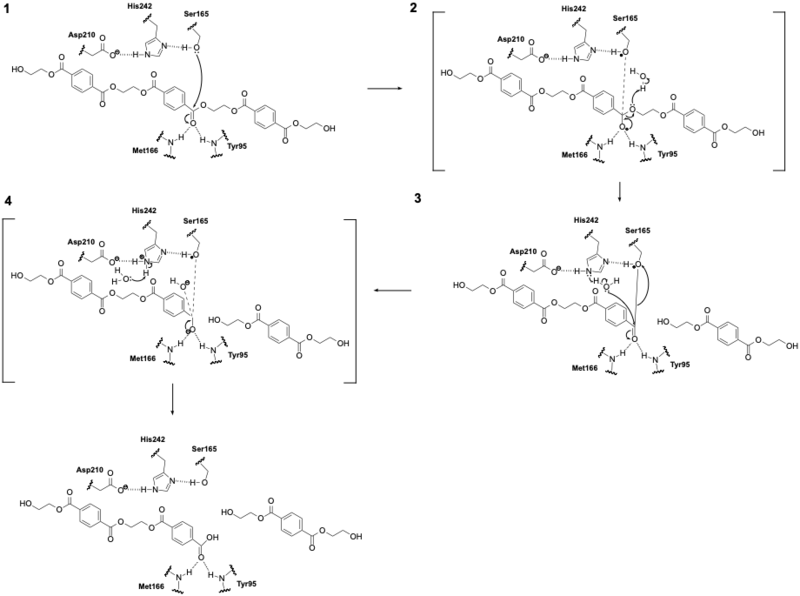
| - | LCC catalyzes the breakdown of PET using a serine hydrolase mechanism with a | + | LCC catalyzes the breakdown of PET using a serine hydrolase mechanism with a <scene name='10/1075247/Catalytic_triad3_w_label/3'>Catalytic Triad</scene> of Ser165, His242, and Asp210. (1) The reaction begins when His242 deprotonates Ser165, which activates it as a nucleophile. (2) Ser165 then attacks the carbonyl carbon of an ester bond in the PET polymer to form a tetrahedral intermediate. (3) This tetrahedral intermediate is stabilized by an oxyanion hole formed by the backbone amides of Met166 and Tyr95. (4) The intermediate collapses; one product is released and an acyl-enzyme intermediate is formed. (5) A water molecule, activated by His242, then attacks the acyl-enzyme. This releases the second product and resets the enzyme’s active site. |
[[Image:Mech2.png|800 px|right|thumb|Figure 1: LCC mechanism. LCC hydrolyzes PET using a catalytic triad (Ser165, His242, Asp210) to cleave ester bonds via a tetrahedral intermediate.]] | [[Image:Mech2.png|800 px|right|thumb|Figure 1: LCC mechanism. LCC hydrolyzes PET using a catalytic triad (Ser165, His242, Asp210) to cleave ester bonds via a tetrahedral intermediate.]] | ||
Revision as of 22:33, 14 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Leaf Branch Compost Cutinase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ 2.0 2.1 2.2 2.3 2.4 Sui B, Wang T, Fang J, Hou Z, Shu T, Lu Z, Liu F, Zhu Y. Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes. Front Microbiol. 2023 Oct 2;14:1265139. PMID:37849919 doi:10.3389/fmicb.2023.1265139
- ↑ Ueda H, Tabata J, Seshime Y, Masaki K, Sameshima-Yamashita Y, Kitamoto H. Cutinase-like biodegradable plastic-degrading enzymes from phylloplane yeasts have cutinase activity. Biosci Biotechnol Biochem. 2021 Jul 23;85(8):1890-1898. PMID:34160605 doi:10.1093/bbb/zbab113
- ↑ Kolattukudy PE. Biopolyester membranes of plants: cutin and suberin. Science. 1980 May 30;208(4447):990-1000. PMID:17779010 doi:10.1126/science.208.4447.990
- ↑ 5.0 5.1 5.2 5.3 5.4 Khairul Anuar NFS, Huyop F, Ur-Rehman G, Abdullah F, Normi YM, Sabullah MK, Abdul Wahab R. An Overview into Polyethylene Terephthalate (PET) Hydrolases and Efforts in Tailoring Enzymes for Improved Plastic Degradation. Int J Mol Sci. 2022 Oct 20;23(20):12644. PMID:36293501 doi:10.3390/ijms232012644
- ↑ 6.0 6.1 Burgin T, Pollard BC, Knott BC, Mayes HB, Crowley MF, McGeehan JE, Beckham GT, Woodcock HL. The reaction mechanism of the Ideonella sakaiensis PETase enzyme. Commun Chem. 2024 Mar 27;7(1):65. PMID:38538850 doi:10.1038/s42004-024-01154-x
- ↑ 7.0 7.1 Stevensen J, Janatunaim RZ, Ratnaputri AH, Aldafa SH, Rudjito RR, Saputro DH, Suhandono S, Putri RM, Aditama R, Fibriani A. Thermostability and Activity Improvements of PETase from Ideonella sakaiensis. ACS Omega. 2025 Feb 12;10(7):6385-6395. PMID:40028137 doi:10.1021/acsomega.4c05142
- ↑ Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, Toyohara K, Miyamoto K, Kimura Y, Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science. 2016 Mar 11;351(6278):1196-9. doi: 10.1126/science.aad6359. PMID:26965627 doi:http://dx.doi.org/10.1126/science.aad6359
- ↑ Landrigan PJ, Stegeman JJ, Fleming LE, Allemand D, Anderson DM, Backer LC, Brucker-Davis F, Chevalier N, Corra L, Czerucka D, Bottein MD, Demeneix B, Depledge M, Deheyn DD, Dorman CJ, Fénichel P, Fisher S, Gaill F, Galgani F, Gaze WH, Giuliano L, Grandjean P, Hahn ME, Hamdoun A, Hess P, Judson B, Laborde A, McGlade J, Mu J, Mustapha A, Neira M, Noble RT, Pedrotti ML, Reddy C, Rocklöv J, Scharler UM, Shanmugam H, Taghian G, van de Water JAJM, Vezzulli L, Weihe P, Zeka A, Raps H, Rampal P. Human Health and Ocean Pollution. Ann Glob Health. 2020 Dec 3;86(1):151. PMID:33354517 doi:10.5334/aogh.2831
- ↑ Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL. Marine pollution. Plastic waste inputs from land into the ocean. Science. 2015 Feb 13;347(6223):768-71. PMID:25678662 doi:10.1126/science.1260352
Student Contributors
Ashley Callaghan Rebecca Hoff Simone McCowan