Introduction
What are Minibinders?
These mini proteins target the interaction between ACE2 and COVID-19 spike protein [1]. The minibinders are small proteins carefully designed to bind to the COVID-19 spike protein with a greater affinity than ACE2 [1]. These minibinders were able to reduce the viral burden of SARS-CoV-2 in mice [2]. These proteins were de novo (from scratch) designs to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein [1]. The binding region between can give a better explanation as to how these proteins were designed.
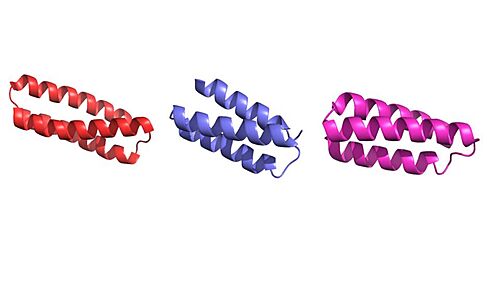
Figure 1. Image of the individual helices of AHB2, LCB1, and LCB3, respectively.
COVID-19 Disease Pathway
Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor, known as ACE2, and this allows the virus to remain anchored to the host for viral entry [3]. When the spike protein binds to the receptor, ACE2 for example, the cell membrane-associated protease, protease serine 2 TMPRSS2 promotes viral entry by activating the spike protein [4]. The activated spike protein is able to cleave itself into S1 and S2 subunits [4]. The S2 subunit is in charge of viral entry and does this through conformational changes [4]. The S2 subunit will insert it's FP domain into the host cell's membrane, and this will trigger an interaction with the HR2 domain and HR1 trimer to form the 6-helical bundle to bring the viral envelope and cell membrane in close enough distance for viral fusion and ultimately viral entry [4]. Once the virus is within the host cell, it is able to translate viral proteins, eliciting an immune response and spreading the viral particles throughout the body [4].
COVID-19 Viral Infection Interruption
The primary goal of the mini binders is to prevent the spike proteins from binding to ACE2, and when the mini binders are bound to the spike protein, the virus is unable to anchor itself to the host protein [1]. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response [1]. Targeting this specific interaction between the COVID-19 spike protein has proven effective and is hopeful target for future therapeutics to treat the virus [4]. LCB1 proved to be quite effective at weakening the immune response, compared to the other mini binders, which can be explained by the binding interface between the spike protein and LCB1 [1].
Expectations of this page
This page will dive into the method of mini binders effectively preventing the entry of the viral SARS-CoV-2 spike protein to enter a host cell. Taking a closer look at the structure of the SARS-CoV-2 spike protein, this will provide a deeper understanding of how it binds to host a cell membrane (ACE2) and give context to how the mini binders are capable of binding to it. Understanding the ACE2 receptor on host cells will give an expectation for how mini binders will bind to the spike protein, stealing the parts of the structure of ACE2 to create a protein with a greater binding affinity to outcompete ACE2.
Laying the foundation for the mini binders, we will then take a look at how the mini binders are designed to obtain the best possible helical structure. With that, then it is finally time to look at the receptor binding domain between the various mini binders and their interactions with the spike protein.
SARS-COV-2 Spike Protein
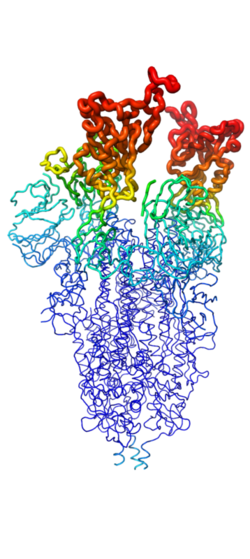
Figure 2. Spike protein shown in "B-Factor"; depicting mobility and flexibility of different portions. Depicted in red are the most mobile, whilst dark blue are the least mobile. The 2 red portions depict RBDs, which correspond to 1-up and 2-up conformational states.
The of SARS-COV-2 is a symmetric trimer featuring 3 spike glycoprotein chains (UNIPROT: P0DTC2). Each monomer of the spike is called a spike glycoprotein, and the total assembly contains 2 main parts: The and subunits[4]. However, the native spike protein does not exist in this state prior to infection. The protein is actually inactive initially, but is later activated by proteases cleaving the inactive S protein into its two active subunits[4]. The S1 subunit contains the . The RBD is responsible for on the surface of the target cell, as well as neutralizing antibodies. The NTD, CTD, and their relevant interfaces actually play much larger roles in the binding of the spike protein to ACE2 than the RBD does due to their larger surface areas[4]. The S2 subunit is responsible for viral fusion and entry. Once bound to ACE2, and after the different domains in S2 have anchored to the membrane as well as delivered the viral envelope, the S2 subunit then changes conformation from the pre-hairpin to conformation[4]. The S2 subunit contains a fusion peptide domain (FP), heptapeptide repeat sequences 1 and 2 (HR1 & HR2), TM domain, and cytoplasmic fusion domain (CT). Full information about the location and structures of these domains within the S2 subunit can be found in references 1 and 3[4][5]. For the purpose of this article about the minibinders, attention will be directed to the S1 subunit and its binding properties with ACE2.
Throughout the entire process, the spike protein has 3 main conformations. An conformation; an active, "open" conformation; and a conformation mentioned previously[4][6][5]. In the closed conformation, the RBDs of each monomer are tucked inwards, preventing interaction. In the open conformation, however, 1 or more of these RBDs can be in the "up" conformation, meaning they are exposed and able to interact within the extracellular space. Mainly, there exits a "" and "" conformation in this phase[6][5]. Depicted in Figure 1, the RBDs of the spike protein have the highest mobility, which further support the many conformational changes in which they are involved. Most of the depictions of the minibinders bound to the spike protein show the spike protein in the 2-up conformation.
ACE2
is a carboxypeptidase present on cell surfaces that is responsible for the degradation of angiotensin II. It is a critical enzyme in the suppression of the renin-angiotensin system. This improves both cardiovascular and renal systems, as well as abates acute respiratory distress syndrome (ARDS). It does the 2 former via the RAS System's role in the regulation of blood pressure, renal function, water homeostasis, electrolyte balance, and/or inflammation[7]. The critical role that this enzyme plays in the regulation of this system is what results in the adverse symptomology observed in victims of the SARS-COV-2 virus. The ACE2 receptor is considered the only essential receptor in the SARS-COV-2 viral mechanism, and thus the collateral debilitation of ACE2 results in the adverse respiratory effects including ARDS, pulmonary edema, destruction of alveolar structures, and others[7]. This relationship was further proven when ACE-2 deficient mice had developed these effects at higher rates compared to the wild type[8].
As mentioned previously, all of the S1 subunit domains play important roles in the . The surface area of the NTD and CTD are particularly important, along with the direct interactions observed in the RBD. Whilst ACE2 is not the focus of this article, understanding its role in the infection pathway of COVID 19, as well as how it binds to the spike protein will assist in understanding the design and functional processes of the minibinders.
Minibinders
Structure
The goal of designing these minibinders was to create a molecule with a higher binding affinity with the RBD than ACE2, meaning they had to be designed with specific residues that form stronger connections with the same binding pockets that ACE2 would bind to [1]. This section highlights some important residue differences between the minibinders and ACE2 that give the minibinders a higher affinity.
The glutamine-493 (Q493) residue is an important residue in showing the differences in strength between ACE2 and the minibinders [1]. ACE2 doesn’t make use of this residue when binding to the RBD, the nearest residues, Glu-35 and Lys-31 . Comparing this to the AHB2 minibinder, which , the AHB2 minibinder makes better use of the RBD’s residue than ACE2, helping it have a higher affinity to the spike protein. LCB1 makes even better use of the Q493 residue, , giving it the highest affinity based on the Q493 residue.
Another important binding site on the RBD includes the Lysine-417 (K417) and Arginine-403 (R403) residues. While using its own D30 residue, LCB1 forms , using its own D30 residue, forming a very strong interaction that is hard to break.
It is important to note that these highlighted residues aren’t the only residues that differ between the minibinders, and it is a compilation of all the residue interactions that give each minibinder different affinities. For example, of the RDB previously mentioned, yet it still has a higher affinity to the RBD than AHB2 which forms a hydrogen bond with the Q493 residue [1].
Function
The effect of the minibinders’ higher affinity to the spike protein RBD is that ACE2’s binding site is now sterically blocked, meaning that it can’t bind to the spike protein and initiate the infection pathway. This effectively hinders the effects of the virus, which was the goal of creating these minibinders[1].
Design
These mini binders, and , were designed from “scratch” (de novo) with the intention to mimic the binding of ACE2 to spike protein [1]. Using Rotamer Interaction Field (RIF) docking, the proteins were able to make the most efficient bonding using the ACE2 spike protein binding interface [1]. Using Site Saturation Mutagenesis (SSM), every residue in the minibinder’s helix scaffold will be substituted with each of the 20 amino acids, one at a time [9]. Forming SSM libraries, each of the libraries converged on a small number of closely related sequences, and from these libraries, the design was selected for AHB2 and LCB1 to find the sequence that yields a protein with a high affinity for the spike proteins receptor binding domain [1]. AHB2 was designed using ACE2 helix scaffold, while LCB1 and LCB3 were designed full from scratch, attempting to make the best possible helix with the greatest affinity for the spike protein receptors [1]. Although LCB1 was designed before LCB3, LCB3 was less effective at neutralizing the viral response with a higher IC50 value [1].
Implications
Potency of Minibinders
Examining the IC50 values of the various minibinders gives quantitative data to the effectiveness of the proteins in preventing an immune response. The highest IC50 was AHB2 (15.5 nM) [1]. The higher IC50 indicates a larger amount of minibinder required to inhibit the biological process. Both LCB1 and LCB3 proved to be significantly more effective than AHB2, “LCB1 and LCB3 were within a factor of 3 of the most potent anti-Spike monoclonal antibodies described to date” [1]. The IC50 values of LCB1 and LCB3 are 23.54 pM and 48.1 pM, respectively[1].
Minibinders in mice
Given that LCB1 proved to be the most effective mini binder, an experiment examined the effectiveness of the mini binder in mice. LCB1 was administered to the mice via nasal delivery. As expected, compared to control mini protein, the LCB1 was significantly more effective at reducing the viral burden, diminishing the immune cell infiltration, and inflammation [2]. The virus was not detected in the lungs 4-7 days post-infection, and the spleen, heart, and brain had viral RNA at very low concentrations [2].
Benefits of minibinders over other therapeutics
The size of these minibinders is a large reason why they are so effective. The minibinders are “5% the molecular weight of a full antibody molecule with… 20-fold more potential for nebulization” [1]. When the LCB1 was modified to attach to a human IgG domain to enhance bioavailability (minibinders stay in the body longer and work more effectively), the results showed that LCB1 was less effective [2]. This can be explained by the proteins being much larger in size. The minibinders are also very stable, so they are able to be administered as a gel via nebulization [1]. Future directions of mini binders are to streamline the process of obtaining a sequence for pathogen neutralizing designs [1]. Given that there are only a small number of antibody therapies and vaccines approved for treatment of COVID, it is important to get this potential therapeutic approved to lay the foundation for other treatments for other viruses. Due to the minibinders being so effective, future directions of mini proteins should look to prevent viral entry of other viruses.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.


