We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
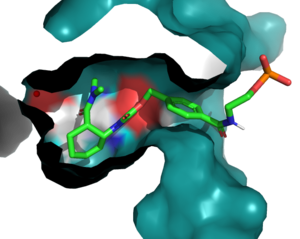
[[Image:BindingPocket.png|300px|right|thumb|Figure 1. Active Site]] | [[Image:BindingPocket.png|300px|right|thumb|Figure 1. Active Site]] | ||
====Active Site==== | ====Active Site==== | ||
| - | In the active state, there are <scene name='10/1075254/Active_site/3'>two catalytic residues</scene> that aim to stabilize the transition state of the Diels-Alder reaction. The Y134 acts as a hydrogen bond donor to the oxygen on the ligand. Q208 acts as a hydrogen bond acceptor to the nitrogen on the ligand. These interactions help reduce the energetic gap between orbitals allowing the reaction to proceed, outlined in HOMO/LUMO. | + | In the active state, there are <scene name='10/1075254/Active_site/3'>two catalytic residues</scene> that aim to stabilize the transition state of the Diels-Alder reaction. The Y134 acts as a hydrogen bond donor to the oxygen on the <scene name='10/1075253/Ligand/6'>ligand</scene>. Q208 acts as a hydrogen bond acceptor to the nitrogen on the ligand. These interactions help reduce the energetic gap between orbitals allowing the reaction to proceed, outlined in HOMO/LUMO. |
====Helix Cap==== | ====Helix Cap==== | ||
In the evolution process, researchers added a 16-residue a-helix motif to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction” (CITE), decreasing the Km of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme. | In the evolution process, researchers added a 16-residue a-helix motif to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction” (CITE), decreasing the Km of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme. | ||
Revision as of 18:26, 15 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Diels-Alderase
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
Taylor Donahue Kate Thuma Micah Zile