User:Milica Nenadovich/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
<scene name='10/1075192/Synf4_main/3'>Syn-F4</scene> is a ''de novo'' ferric enterobactin esterase designed to highlight the potential of synthetic biology in creating simpler protein structures capable of performing complex functions <ref name="Kurihara">PMID:37695900</ref>. Syn-F4 is also the first ''de novo'' protein catalytically active ''in vitro'' and biologically functional ''in vivo'' <ref name="Kurihara"/>. Ferric enterobactin esterases are a type of endogenous bacterial enzyme that break ester bonds of small metabolites bound to iron, allowing iron to be released into and utilized by bacterial cells <ref name="Abergel">PMID:16819888</ref> <ref name="Peuckert">PMID:21802011</ref>. Iron plays crucial roles in various physiological processes in bacteria, including cellular respiration, oxidative stress responses, and DNA and protein synthesis <ref name="Peuckert"/>. | <scene name='10/1075192/Synf4_main/3'>Syn-F4</scene> is a ''de novo'' ferric enterobactin esterase designed to highlight the potential of synthetic biology in creating simpler protein structures capable of performing complex functions <ref name="Kurihara">PMID:37695900</ref>. Syn-F4 is also the first ''de novo'' protein catalytically active ''in vitro'' and biologically functional ''in vivo'' <ref name="Kurihara"/>. Ferric enterobactin esterases are a type of endogenous bacterial enzyme that break ester bonds of small metabolites bound to iron, allowing iron to be released into and utilized by bacterial cells <ref name="Abergel">PMID:16819888</ref> <ref name="Peuckert">PMID:21802011</ref>. Iron plays crucial roles in various physiological processes in bacteria, including cellular respiration, oxidative stress responses, and DNA and protein synthesis <ref name="Peuckert"/>. | ||
| - | |||
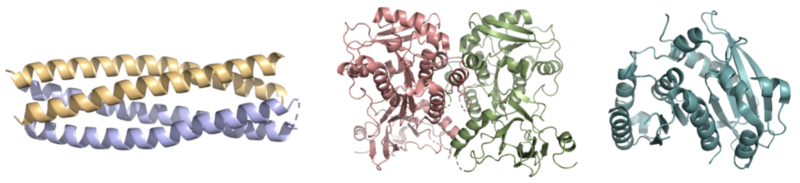
| - | [[Image: FesCompare.png|500 px|center|thumb|'''Figure 1.''' Pictured left-to-right: Structures of the ferric enterobactin esterases Syn-F4 (PDB: 8H7C), Fes (PDB: 3C87), and IroE (PDB: 2GZR), respectively.]] | ||
<scene name='10/1075192/Fes/3'>Fes</scene> is one such native esterase responsible for releasing Fe<sup>3+</sup> from iron-chelating compounds, such as [https://en.wikipedia.org/wiki/Enterobactin ferric enterobactin], within the cell, making it critical for bacterial survival <ref name="Abergel"/> <ref name="Lin">PMID:16076215</ref>. In order to mimic evolutionary selection of proteins for specific biological functions, Syn-IF was a ''de novo'' bifunctional protein designed to perform iron release, similar to Fes. Syn-IF was subjected to a series of random mutagenesis to individually select for its iron-releasing function, leading to the creation of Syn-F4 <ref name="Smith">PMID:25420677</ref>. While Syn-F4 catalysis is 1000-fold slower than that of a native Fes enzyme, it is still sufficient in rescuing function in ''∆fes E.coli'' <ref name="Kurihara"/> <ref name="Smith"/>. | <scene name='10/1075192/Fes/3'>Fes</scene> is one such native esterase responsible for releasing Fe<sup>3+</sup> from iron-chelating compounds, such as [https://en.wikipedia.org/wiki/Enterobactin ferric enterobactin], within the cell, making it critical for bacterial survival <ref name="Abergel"/> <ref name="Lin">PMID:16076215</ref>. In order to mimic evolutionary selection of proteins for specific biological functions, Syn-IF was a ''de novo'' bifunctional protein designed to perform iron release, similar to Fes. Syn-IF was subjected to a series of random mutagenesis to individually select for its iron-releasing function, leading to the creation of Syn-F4 <ref name="Smith">PMID:25420677</ref>. While Syn-F4 catalysis is 1000-fold slower than that of a native Fes enzyme, it is still sufficient in rescuing function in ''∆fes E.coli'' <ref name="Kurihara"/> <ref name="Smith"/>. | ||
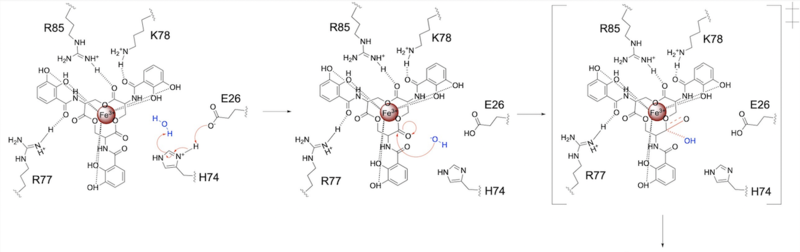
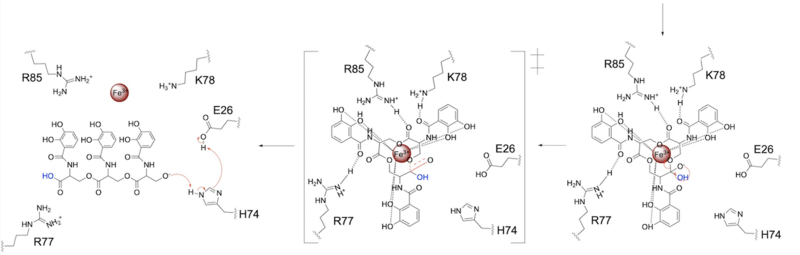
Whereas most ferric enterobactin esterases are [https://en.wikipedia.org/wiki/Serine_hydrolase serine hydrolases] that function via a catalytic triad, such as Fes, Syn-F4 performs its function via a catalytic dyad without the involvement of a serine active site residue ('''Fig. 1''') <ref name="Kurihara"/> <ref name="Lin">PMID:16076215</ref>. One other serine hydrolase, <scene name='10/1075192/Iroe/2'>IroE</scene>, also performs catalysis through a catalytic dyad but still uses a serine residue as the nucleophile ('''Fig. 1''') <ref name="Lin">PMID:16076215</ref> <ref name="Larsen">PMID:16922493</ref>. The unassuming design of Syn-F4 was modeled after the ''de novo'' protein <scene name='10/1075192/Wa20/3'>WA20</scene>, which was originally designed to be used as a "building block" for other synthetic proteins and nanotechnology designs <ref name="Arai">PMID:22397676</ref>. The crystallization of WA20 reveals an alpha-helix bundle structure, which served as the basis for the design of Syn-F4 <ref name="Kurihara"/> <ref name="Arai">PMID:22397676</ref> . | Whereas most ferric enterobactin esterases are [https://en.wikipedia.org/wiki/Serine_hydrolase serine hydrolases] that function via a catalytic triad, such as Fes, Syn-F4 performs its function via a catalytic dyad without the involvement of a serine active site residue ('''Fig. 1''') <ref name="Kurihara"/> <ref name="Lin">PMID:16076215</ref>. One other serine hydrolase, <scene name='10/1075192/Iroe/2'>IroE</scene>, also performs catalysis through a catalytic dyad but still uses a serine residue as the nucleophile ('''Fig. 1''') <ref name="Lin">PMID:16076215</ref> <ref name="Larsen">PMID:16922493</ref>. The unassuming design of Syn-F4 was modeled after the ''de novo'' protein <scene name='10/1075192/Wa20/3'>WA20</scene>, which was originally designed to be used as a "building block" for other synthetic proteins and nanotechnology designs <ref name="Arai">PMID:22397676</ref>. The crystallization of WA20 reveals an alpha-helix bundle structure, which served as the basis for the design of Syn-F4 <ref name="Kurihara"/> <ref name="Arai">PMID:22397676</ref> . | ||
| + | |||
| + | [[Image: FesCompare.png|800 px|center|thumb|'''Figure 1.''' Pictured left-to-right: Structures of the ferric enterobactin esterases Syn-F4 (PDB: 8H7C), Fes (PDB: 3C87), and IroE (PDB: 2GZR), respectively.]] | ||
====Ligand==== | ====Ligand==== | ||
Revision as of 14:29, 22 April 2025
Syn-F4, a de novo Ferric Enterobactin Esterase
| |||||||||||
Student Contributors
- Milica Nenadovich
- Reesha Bhagat