User:Hayden Vissing/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
Plastic pollution is a major problem in our modern world. Many strategies have been employed to recycle plastics instead of creating more and adding further pollution to the environment. | Plastic pollution is a major problem in our modern world. Many strategies have been employed to recycle plastics instead of creating more and adding further pollution to the environment. | ||
| - | One of the most commonly used plastics is a polymer called Polyethylene Terephthalate, also known as [https://en.wikipedia.org/wiki/Polyethylene_terephthalate PET]. PET is commonly used to make plastic bottles or food containers due to its thermal stability, transparency, and impermeability to liquids and gases. Unfortunately, these properties that make it desirable also make it non-biodegradable, meaning that we must recycle PET to avoid polluting the environment with it. Traditional methods, such as melting and reforming the plastic, destroy the mechanical properties of PET, eliminating the ability to recycle it and necessitating a new strategy for recycling PET. | + | One of the most commonly used plastics is a polymer called Polyethylene Terephthalate, also known as [https://en.wikipedia.org/wiki/Polyethylene_terephthalate PET]. PET is commonly used to make plastic bottles or food containers due to its thermal stability, transparency, and impermeability to liquids and gases. Unfortunately, these properties that make it desirable also make it non-biodegradable, meaning that we must recycle PET to avoid polluting the environment with it. Traditional methods, such as melting and reforming the plastic, destroy the mechanical properties of PET, eliminating the ability to recycle it and necessitating a new strategy for recycling PET<ref name="biodegradation">Hiraga, K., Taniguchi, I., Yoshida, S., Kimura, Y., & Oda, K. (2019). Biodegradation of waste PET: A sustainable solution for dealing with plastic pollution. EMBO Reports, 20(11), e49365. https://doi.org/10.15252/embr.201949365. [Published correction appears in EMBO Reports, 21(2), e49826. [https://doi.org/10.15252/embr.201949826. DOI: 10.15252/embr.201949826]</ref>. |
===Enzyme hydrolysis=== | ===Enzyme hydrolysis=== | ||
| - | One solution that researchers have been studying is using a naturally occurring enzyme that has PET degrading activity, PET hyrdolase, to break down the polymer into its monomers<ref> | + | One solution that researchers have been studying is using a naturally occurring enzyme that has PET degrading activity, PET hyrdolase, to break down the polymer into its monomers<ref>Jayasekara, S. K., Joni, H. D., Jayantha, B., Dissanayake, L., Mandrell, C., Sinharage, M. M. S., Molitor, R., Jayasekara, T., Sivakumar, P., & Jayakody, L. N. (2023). Trends in in-silico guided engineering of efficient polyethylene terephthalate (PET) hydrolyzing enzymes to enable bio-recycling and upcycling of PET. Computational and structural biotechnology journal, 21, 3513–3521. [https://doi.org/10.1016/j.csbj.2023.06.004 DOI: 10.1016/j.csbj.2023.06.004]</ref>. This would allow the PET to be reformed from its monomers into a new piece of plastic. |
A class of enzymes that has been a focus is [https://en.wikipedia.org/wiki/Cutinase Cutinase]. | A class of enzymes that has been a focus is [https://en.wikipedia.org/wiki/Cutinase Cutinase]. | ||
| Line 15: | Line 15: | ||
===Leaf-branch compost cutinase (LCC)=== | ===Leaf-branch compost cutinase (LCC)=== | ||
| - | The study done by Tournier et al. (2020)<ref>PMID:32269349</ref> identified a good template enzyme for creating a PET hydrolase as the LCC enzyme. This enzyme was discovered through [https://en.wikipedia.org/wiki/Metagenomics metagenomics] of a leaf-branch compost. Its target substrate is the polymer [https://en.wikipedia.org/wiki/Cutin cutin], found in the cuticle of plants. LCC breaks down Cutin to separate the monomers at the ester bond between them. The LCC enzyme was compared to several other enzymes and was selected as it showed the best rate of PET depolymerization | + | The study done by Tournier et al. (2020)<ref>PMID:32269349</ref> identified a good template enzyme for creating a PET hydrolase as the LCC enzyme. This enzyme was discovered through [https://en.wikipedia.org/wiki/Metagenomics metagenomics] of a leaf-branch compost. Its target substrate is the polymer [https://en.wikipedia.org/wiki/Cutin cutin], found in the cuticle of plants. LCC breaks down Cutin to separate the monomers at the ester bond between them<ref name="breakdown">PMID:29374183</ref>. The LCC enzyme was compared to several other enzymes and was selected as it showed the best rate of PET depolymerization<ref>PMID:22194294</ref>. |
==Mechanism== | ==Mechanism== | ||
===Overview of Active Site=== | ===Overview of Active Site=== | ||
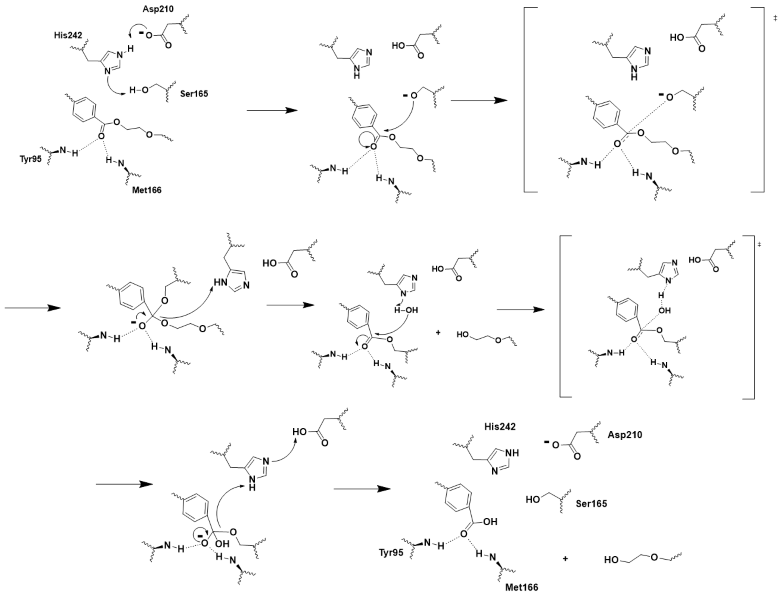
| - | LCC is a serine hydrolase. The mechanism uses a <scene name='10/1076051/Wt_cat_triad/3'>catalytic triad</scene>: S165 (which was made into an alanine for crystallization purposes), D210, and H242. S165 is the nucleophile attacking the electrophilic carbonyl carbon with the <scene name='10/1075218/Oxyanion_hole/4'>oxyanion hole</scene> to stabilize the translation state. These key residues of the catalytic triad and oxyanion hole help to make up the <scene name='10/1076051/Wt_active_site/1'>active site</scene><ref>PMID:32269349</ref>. | + | LCC is a serine hydrolase. The mechanism uses a <scene name='10/1076051/Wt_cat_triad/3'>catalytic triad</scene>: S165 (which was made into an alanine for crystallization purposes), D210, and H242. S165 is the nucleophile attacking the electrophilic carbonyl carbon with the <scene name='10/1075218/Oxyanion_hole/4'>oxyanion hole</scene> to stabilize the translation state<ref>Han, X., Liu, W., Huang, J. W., et al. (2017). Structural insight into catalytic mechanism of PET hydrolase. Nature Communications, 8, 2106. https://doi.org/10.1038/s41467-017-02255-z.Heredia-Guerrero, J. A., Heredia, A., García-Segura, R., & Benítez, J. J. (2009). Synthesis and characterization of a plant cutin mimetic polymer. Polymer, 50(24), 5633–5637. [https://doi.org/10.1016/j.polymer.2009.10.018 DOI: 10.1016/j.polymer.2009.10.018]</ref>. These key residues of the catalytic triad and oxyanion hole help to make up the <scene name='10/1076051/Wt_active_site/1'>active site</scene><ref>PMID:32269349</ref>. |
[[Image:Push_mechanism.png|800px|center|thumb|Figure 2 - Push-mechanism for the cutinase reaction]] | [[Image:Push_mechanism.png|800px|center|thumb|Figure 2 - Push-mechanism for the cutinase reaction]] | ||
Revision as of 18:34, 22 April 2025
The Future of Recycling: the ICCG Mutant Pet Hydroalase
| |||||||||||
References
- ↑ Hiraga, K., Taniguchi, I., Yoshida, S., Kimura, Y., & Oda, K. (2019). Biodegradation of waste PET: A sustainable solution for dealing with plastic pollution. EMBO Reports, 20(11), e49365. https://doi.org/10.15252/embr.201949365. [Published correction appears in EMBO Reports, 21(2), e49826. DOI: 10.15252/embr.201949826
- ↑ Jayasekara, S. K., Joni, H. D., Jayantha, B., Dissanayake, L., Mandrell, C., Sinharage, M. M. S., Molitor, R., Jayasekara, T., Sivakumar, P., & Jayakody, L. N. (2023). Trends in in-silico guided engineering of efficient polyethylene terephthalate (PET) hydrolyzing enzymes to enable bio-recycling and upcycling of PET. Computational and structural biotechnology journal, 21, 3513–3521. DOI: 10.1016/j.csbj.2023.06.004
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, Choi SY, Lee SY, Kim KJ. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat Commun. 2018 Jan 26;9(1):382. doi: 10.1038/s41467-018-02881-1. PMID:29374183 doi:http://dx.doi.org/10.1038/s41467-018-02881-1
- ↑ Sulaiman S, Yamato S, Kanaya E, Kim JJ, Koga Y, Takano K, Kanaya S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol. 2012 Mar;78(5):1556-62. doi: 10.1128/AEM.06725-11. Epub, 2011 Dec 22. PMID:22194294 doi:http://dx.doi.org/10.1128/AEM.06725-11
- ↑ Han, X., Liu, W., Huang, J. W., et al. (2017). Structural insight into catalytic mechanism of PET hydrolase. Nature Communications, 8, 2106. https://doi.org/10.1038/s41467-017-02255-z.Heredia-Guerrero, J. A., Heredia, A., García-Segura, R., & Benítez, J. J. (2009). Synthesis and characterization of a plant cutin mimetic polymer. Polymer, 50(24), 5633–5637. DOI: 10.1016/j.polymer.2009.10.018
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
- ↑ Tournier V, Topham CM, Gilles A, David B, Folgoas C, Moya-Leclair E, Kamionka E, Desrousseaux ML, Texier H, Gavalda S, Cot M, Guemard E, Dalibey M, Nomme J, Cioci G, Barbe S, Chateau M, Andre I, Duquesne S, Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020 Apr;580(7802):216-219. doi: 10.1038/s41586-020-2149-4. Epub 2020 Apr, 8. PMID:32269349 doi:http://dx.doi.org/10.1038/s41586-020-2149-4
Student Contributors
- David Bogle
- Justin Chavez
- Hayden Vissing