User:Reesha Bhagat/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 63: | Line 63: | ||
''De novo'' proteins are at the forefront of biochemical and industrial applications, designed to mimic the evolutionary selection of proteins for highly specific functions. These engineered proteins show significant promise in enhancing antibiotic therapies, particularly in light of rising [https://en.wikipedia.org/wiki/Antimicrobial_resistance antibiotic resistance]. For example, targeting bacterial iron acquisition pathways may offer an effective alternative to traditional antibiotic treatments <ref name="Stelitano">PMID:37047161</ref> <ref name="Rivera">PMID:37451083</ref>. Furthermore, as our understanding of cellular signaling pathways advances, ''de novo'' proteins present new opportunities for cancer therapy. Through [https://en.wikipedia.org/wiki/Synthetic_biology synthetic biology], it is possible to increase drug efficacy by designing agents that selectively target malignant cells while preserving healthy tissue <ref name="Lim">PMID:35255684</ref>. Additionally, programming bacteria through synthetic compounds could enable bacteriotherapy approaches, in which engineered bacteria express specific proteins that bind to tumor-associated antigen markers, offering a novel strategy for cancer targeting and treatment <ref name="Mansour">PMID:30950210</ref>. Synthetic biology holds vast potential across a multitude of fields, with applications that span far beyond initial application. Its development is set to become integral to the future of biomedical research and innovation. As the field of synthetic biology continues to advance, its potential applications appear boundless <ref name="Thevenin">PMID:9354688</ref> <ref name="Yan">PMID:37169742</ref>. | ''De novo'' proteins are at the forefront of biochemical and industrial applications, designed to mimic the evolutionary selection of proteins for highly specific functions. These engineered proteins show significant promise in enhancing antibiotic therapies, particularly in light of rising [https://en.wikipedia.org/wiki/Antimicrobial_resistance antibiotic resistance]. For example, targeting bacterial iron acquisition pathways may offer an effective alternative to traditional antibiotic treatments <ref name="Stelitano">PMID:37047161</ref> <ref name="Rivera">PMID:37451083</ref>. Furthermore, as our understanding of cellular signaling pathways advances, ''de novo'' proteins present new opportunities for cancer therapy. Through [https://en.wikipedia.org/wiki/Synthetic_biology synthetic biology], it is possible to increase drug efficacy by designing agents that selectively target malignant cells while preserving healthy tissue <ref name="Lim">PMID:35255684</ref>. Additionally, programming bacteria through synthetic compounds could enable bacteriotherapy approaches, in which engineered bacteria express specific proteins that bind to tumor-associated antigen markers, offering a novel strategy for cancer targeting and treatment <ref name="Mansour">PMID:30950210</ref>. Synthetic biology holds vast potential across a multitude of fields, with applications that span far beyond initial application. Its development is set to become integral to the future of biomedical research and innovation. As the field of synthetic biology continues to advance, its potential applications appear boundless <ref name="Thevenin">PMID:9354688</ref> <ref name="Yan">PMID:37169742</ref>. | ||
| - | |||
</StructureSection> | </StructureSection> | ||
| Line 70: | Line 69: | ||
==Student Contributors== | ==Student Contributors== | ||
| + | *Milica Nenadovich | ||
*Reesha Bhagat | *Reesha Bhagat | ||
| - | *Milica Nenadovich | ||
Revision as of 13:45, 23 April 2025
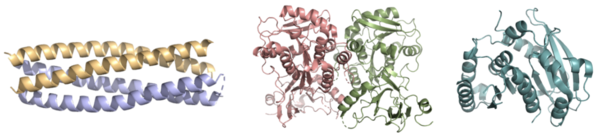
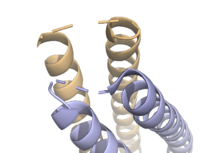
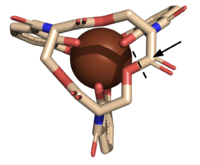
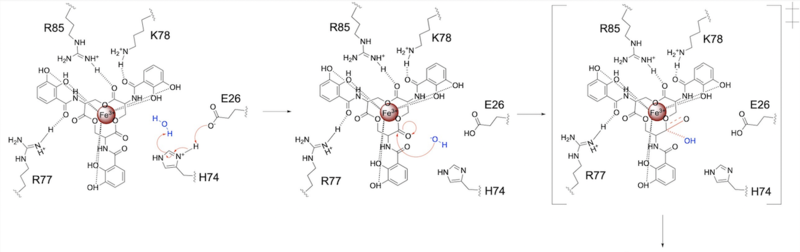
Syn-F4, a de novo Ferric Enterobactin Esterase
| |||||||||||
References
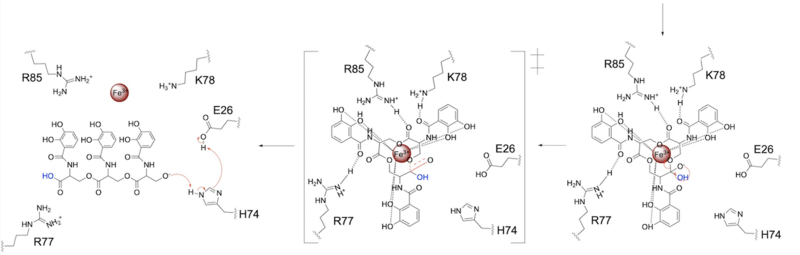
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Kurihara K, Umezawa K, Donnelly AE, Sperling B, Liao G, Hecht MH, Arai R. Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4. Proc Natl Acad Sci U S A. 2023 Sep 19;120(38):e2218281120. PMID:37695900 doi:10.1073/pnas.2218281120
- ↑ 2.0 2.1 2.2 2.3 2.4 Abergel RJ, Warner JA, Shuh DK, Raymond KN. Enterobactin protonation and iron release: structural characterization of the salicylate coordination shift in ferric enterobactin. J Am Chem Soc. 2006 Jul 12;128(27):8920-31. PMID:16819888 doi:10.1021/ja062046j
- ↑ 3.0 3.1 3.2 Peuckert F, Ramos-Vega AL, Miethke M, Schworer CJ, Albrecht AG, Oberthur M, Marahiel MA. The siderophore binding protein FeuA shows limited promiscuity toward exogenous triscatecholates. Chem Biol. 2011 Jul 29;18(7):907-19. PMID:21802011 doi:10.1016/j.chembiol.2011.05.006
- ↑ 4.0 4.1 4.2 4.3 4.4 Lin H, Fischbach MA, Liu DR, Walsh CT. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J Am Chem Soc. 2005 Aug 10;127(31):11075-84. PMID:16076215 doi:10.1021/ja0522027
- ↑ 5.0 5.1 Smith BA, Mularz AE, Hecht MH. Divergent evolution of a bifunctional de novo protein. Protein Sci. 2015 Feb;24(2):246-52. PMID:25420677 doi:10.1002/pro.2611
- ↑ 6.0 6.1 6.2 Larsen NA, Lin H, Wei R, Fischbach MA, Walsh CT. Structural characterization of enterobactin hydrolase IroE. Biochemistry. 2006 Aug 29;45(34):10184-90. PMID:16922493 doi:10.1021/bi060950i
- ↑ 7.0 7.1 7.2 Arai R, Kobayashi N, Kimura A, Sato T, Matsuo K, Wang AF, Platt JM, Bradley LH, Hecht MH. Domain-Swapped Dimeric Structure of a Stable and Functional De Novo 4-Helix Bundle Protein, WA20. J Phys Chem B. 2012 Mar 8. PMID:22397676 doi:10.1021/jp212438h
- ↑ 8.0 8.1 Moynie L, Milenkovic S, Mislin GLA, Gasser V, Malloci G, Baco E, McCaughan RP, Page MGP, Schalk IJ, Ceccarelli M, Naismith JH. The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat Commun. 2019 Aug 14;10(1):3673. doi: 10.1038/s41467-019-11508-y. PMID:31413254 doi:http://dx.doi.org/10.1038/s41467-019-11508-y
- ↑ 9.0 9.1 Timofeeva AM, Galyamova MR, Sedykh SE. Bacterial Siderophores: Classification, Biosynthesis, Perspectives of Use in Agriculture. Plants (Basel). 2022 Nov 12;11(22):3065. PMID:36432794 doi:10.3390/plants11223065
- ↑ Stelitano G, Cocorullo M, Mori M, Villa S, Meneghetti F, Chiarelli LR. Iron Acquisition and Metabolism as a Promising Target for Antimicrobials (Bottlenecks and Opportunities): Where Do We Stand? Int J Mol Sci. 2023 Mar 24;24(7):6181. PMID:37047161 doi:10.3390/ijms24076181
- ↑ Rivera M. Mobilization of iron stored in bacterioferritin, a new target for perturbing iron homeostasis and developing antibacterial and antibiofilm molecules. J Inorg Biochem. 2023 Oct;247:112306. PMID:37451083 doi:10.1016/j.jinorgbio.2023.112306
- ↑ Lim B, Yin Y, Ye H, Cui Z, Papachristodoulou A, Huang WE. Reprogramming Synthetic Cells for Targeted Cancer Therapy. ACS Synth Biol. 2022 Mar 18;11(3):1349-1360. PMID:35255684 doi:10.1021/acssynbio.1c00631
- ↑ Sedighi M, Zahedi Bialvaei A, Hamblin MR, Ohadi E, Asadi A, Halajzadeh M, Lohrasbi V, Mohammadzadeh N, Amiriani T, Krutova M, Amini A, Kouhsari E. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019 Jun;8(6):3167-3181. PMID:30950210 doi:10.1002/cam4.2148
- ↑ Thevenin BJ, Crandall I, Ballas SK, Sherman IW, Shohet SB. Band 3 peptides block the adherence of sickle cells to endothelial cells in vitro. Blood. 1997 Nov 15;90(10):4172-9 PMID:9354688
- ↑ Yan X, Liu X, Zhao C, Chen GQ. Applications of synthetic biology in medical and pharmaceutical fields. Signal Transduct Target Ther. 2023 May 11;8(1):199. PMID:37169742 doi:10.1038/s41392-023-01440-5
Student Contributors
- Milica Nenadovich
- Reesha Bhagat