User:Reesha Bhagat/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
====Alpha-Helix Bundle==== | ====Alpha-Helix Bundle==== | ||
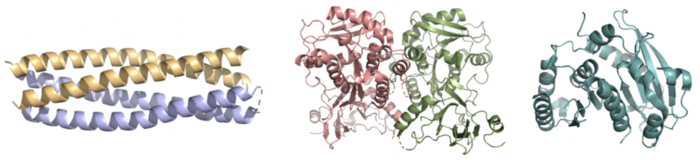
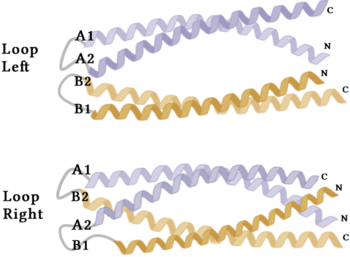
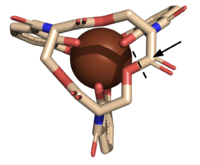
| - | The crystallization of the ''de novo'' protein, WA20, revealed | + | The crystallization of the ''de novo'' protein, WA20, revealed an <scene name='10/1075192/Overlay_rotate/3'>alpha-helix bundle</scene> homodimer structure, which served as the foundation for the design of Syn-F4 <ref name="Arai"/>. While structurally similar, Syn-F4 features a novel active site characterized by the presence of a <scene name='10/1075192/Central_hole_lone/2'>central hole</scene> <ref name="Kurihara"/>. [[Image:LOOOOPS.png|350 px|left|thumb|'''Figure 4.''' A schematic representing the differential conformations of the "loop-left" and "loop-right" Syn-F4 structures (Image created in BioRender.com).]] The symmetry of the four-helix bundle, combined with the configuration of the central hole, facilitates catalytic activity by effectively cleaving the ester bond in ferric enterobactin <ref name="Kurihara"/>. |
====Stabilizing Factors==== | ====Stabilizing Factors==== | ||
| Line 31: | Line 31: | ||
The <scene name='10/1075192/Terminal_residues/3'>terminal residues</scene> on either side of the bundles largely contribute to the stability of Syn-F4. Due to the symmetry of the chains, both '''{{Font color|#a899e6|chain A}}''' and '''{{Font color|#d6b588|chain B}}''' display the same residues and are linked by unresolved residues ('''Fig. 3''') <ref name="Kurihara"/>. The accumulated negative charge on the C-termini is stabilized by positively-charged residues (H10, H45, H46, R102), and conversely, the accumulated positive charge on the N-termini is stabilized by negatively-charged residues (D59). Additionally, the <scene name='10/1075192/H2ophobic/2'>hydrophobic residues</scene> within the core of Syn-F4 interact via Van der Waals to promote association between the helices. | The <scene name='10/1075192/Terminal_residues/3'>terminal residues</scene> on either side of the bundles largely contribute to the stability of Syn-F4. Due to the symmetry of the chains, both '''{{Font color|#a899e6|chain A}}''' and '''{{Font color|#d6b588|chain B}}''' display the same residues and are linked by unresolved residues ('''Fig. 3''') <ref name="Kurihara"/>. The accumulated negative charge on the C-termini is stabilized by positively-charged residues (H10, H45, H46, R102), and conversely, the accumulated positive charge on the N-termini is stabilized by negatively-charged residues (D59). Additionally, the <scene name='10/1075192/H2ophobic/2'>hydrophobic residues</scene> within the core of Syn-F4 interact via Van der Waals to promote association between the helices. | ||
| - | The orientation of the alpha helices and their associated interactions could not be definitively determined due to the absence of loop residues in the crystallized structure of Syn-F4 <ref name="Kurihara"/>. To address this, molecular dynamics (MD) simulations were performed to estimate the predominant conformation of the helices <ref name="Kurihara"/>. Two potential conformations were identified: "loop left" and "loop right" ('''Fig. 4''') <ref name="Kurihara"/>. Among these, the "loop left" conformation demonstrated greater stability and was therefore considered the preferred structure ('''Fig. 4''') <ref name="Kurihara"/>. | + | The orientation of the alpha helices and their associated interactions could not be definitively determined due to the absence of loop residues in the crystallized structure of Syn-F4 ('''Fig. 3''') <ref name="Kurihara"/>. To address this, molecular dynamics (MD) simulations were performed to estimate the predominant conformation of the helices <ref name="Kurihara"/>. Two potential conformations were identified: "loop left" and "loop right" ('''Fig. 4''') <ref name="Kurihara"/>. Among these, the "loop left" conformation demonstrated greater stability and was therefore considered the preferred structure ('''Fig. 4''') <ref name="Kurihara"/>. |
== Function == | == Function == | ||
| Line 47: | Line 47: | ||
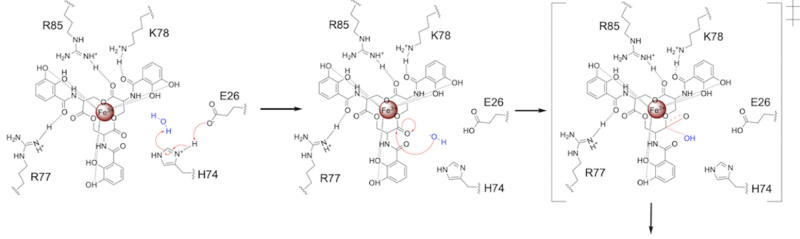
=====Step 1===== | =====Step 1===== | ||
| - | E26 first deprotonates H74, which then deprotonates | + | E26 first deprotonates H74, which then deprotonates H<sub>2</sub>O in turn ('''Fig. 6'''). The newly-formed hydroxide attacks the ester electrophile, kicking electron density up onto the carboxyl group in the first transition state ('''Fig. 6'''). |
[[Image: SynMech1.png|800 px|center|thumb|'''Figure 6.''' Step 1. Activation of water as the nucleophile and attack of the ester electrophile. The first transition state is depicted.]] | [[Image: SynMech1.png|800 px|center|thumb|'''Figure 6.''' Step 1. Activation of water as the nucleophile and attack of the ester electrophile. The first transition state is depicted.]] | ||
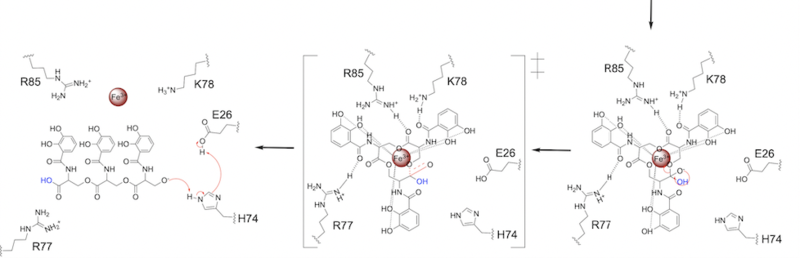
| Line 57: | Line 57: | ||
[[Image: SynMech22222.png|800 px|center|thumb|'''Figure 7.''' Step 2. The second transition state and linearization of ferric enterobactin are depicted. Iron is released and the enzyme resets itself.]] | [[Image: SynMech22222.png|800 px|center|thumb|'''Figure 7.''' Step 2. The second transition state and linearization of ferric enterobactin are depicted. Iron is released and the enzyme resets itself.]] | ||
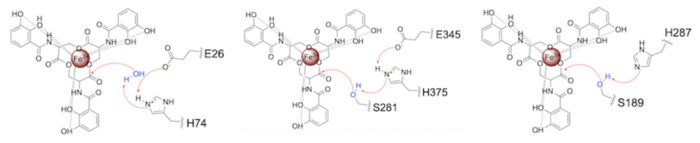
| - | ====Related | + | ====Related Enzymes==== |
Both the structure and mechanism of Syn-F4 differ from other native ferric enterobactin esterases whose structures have been previously determined. Native Fes and IroE enzymes are both serine hydrolases, which means a serine residue acts as their nucleophile <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva">PMID:36432794</ref>. Native <scene name='10/1075192/Fes_residues/11'>Fes</scene> is a homodimer that acts via a catalytic triad (S281, E345, and H375), whereas <scene name='10/1075192/Iroe_residue/6'>IroE</scene> acts via a catalytic dyad (S189 and H287), similar to Syn-F4 ('''Fig. 8''') <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva"/>. | Both the structure and mechanism of Syn-F4 differ from other native ferric enterobactin esterases whose structures have been previously determined. Native Fes and IroE enzymes are both serine hydrolases, which means a serine residue acts as their nucleophile <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva">PMID:36432794</ref>. Native <scene name='10/1075192/Fes_residues/11'>Fes</scene> is a homodimer that acts via a catalytic triad (S281, E345, and H375), whereas <scene name='10/1075192/Iroe_residue/6'>IroE</scene> acts via a catalytic dyad (S189 and H287), similar to Syn-F4 ('''Fig. 8''') <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva"/>. | ||
| Line 74: | Line 74: | ||
==Student Contributors== | ==Student Contributors== | ||
| - | *Milica Nenadovich | ||
*Reesha Bhagat | *Reesha Bhagat | ||
| + | *Milica Nenadovich | ||
Revision as of 12:26, 25 April 2025
Syn-F4, a de novo Ferric Enterobactin Esterase
| |||||||||||
Student Contributors
- Reesha Bhagat
- Milica Nenadovich