We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Carson Powers/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
=''De novo miniprotein Covid therapeutic''= | =''De novo miniprotein Covid therapeutic''= | ||
<StructureSection load='7jzl' size='350' side='right' caption='LCB1 bound to spike protein. LCB1 is colored pink and shown in cartoon. The spike protein is colored by chain and shown by its surface. PDB file: 7jzl' scene='10/1076041/Spike_with_lcb1/3'> | <StructureSection load='7jzl' size='350' side='right' caption='LCB1 bound to spike protein. LCB1 is colored pink and shown in cartoon. The spike protein is colored by chain and shown by its surface. PDB file: 7jzl' scene='10/1076041/Spike_with_lcb1/3'> | ||
| - | |||
| - | |||
=Introduction= | =Introduction= | ||
| - | |||
| - | |||
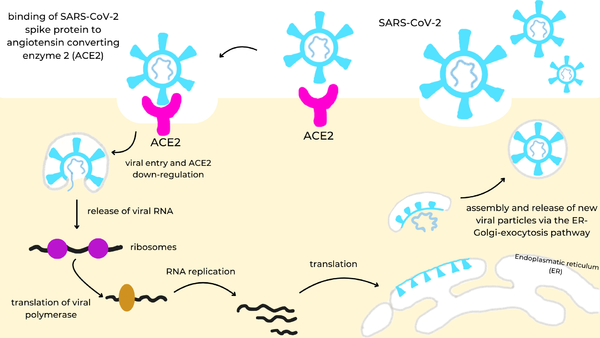
SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA virus that is responsible for the highly infectious respiratory disease, COVID-19.[[Image:spike_infection_cycle.png|600px|right|SARS-CoV-2 infection cycle: the viral spike protein (cyan) binds to the ACE2 receptor (magenta) on the cell surface, triggering endocytosis and ACE2 down-regulation, release of viral RNA into the cytosol, translation of the viral polymerase and genome replication, synthesis of structural proteins in the endoplasmic reticulum (ER), and assembly and release of new virions via the ER-Golgi-exocytosis pathway.]] | SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA virus that is responsible for the highly infectious respiratory disease, COVID-19.[[Image:spike_infection_cycle.png|600px|right|SARS-CoV-2 infection cycle: the viral spike protein (cyan) binds to the ACE2 receptor (magenta) on the cell surface, triggering endocytosis and ACE2 down-regulation, release of viral RNA into the cytosol, translation of the viral polymerase and genome replication, synthesis of structural proteins in the endoplasmic reticulum (ER), and assembly and release of new virions via the ER-Golgi-exocytosis pathway.]] | ||
==Overall SARS-CoV-2 Spike Structure== | ==Overall SARS-CoV-2 Spike Structure== | ||
| - | It is made up of four structural proteins: spike, envelope, membrane, and nucleocapsid proteins. The spike protein helps the virus infect human cells. The envelope protein helps in virus assembly and release. The membrane protein shapes the viral envelope. The nucleocapsid protein binds the RNA genome for replication. Once inside the cell, virus-specific RNA and proteins are synthesized within the cytoplasm. The viral envelope merges with the oily membrane of our own cells, allowing the virus to release its genetic material into the inside of the healthy cell. The genetic blueprint of the virus is RNA instead of DNA so it gives infected host cells instructions to read the template and translate it to replicate the virus. Each infected cell may produce and release millions of copies of the virus which can infect other neighboring cells and people when the viral particles are released from the airways (i.e., via coughing or sneezing). | + | It is made up of four structural proteins: spike, envelope, membrane, and nucleocapsid proteins. The spike protein helps the virus infect human cells. The envelope protein helps in virus assembly and release. The membrane protein shapes the viral envelope. The nucleocapsid protein binds the RNA genome for replication. Once inside the cell, virus-specific RNA and proteins are synthesized within the cytoplasm. The viral envelope merges with the oily membrane of our own cells, allowing the virus to release its genetic material into the inside of the healthy cell. The genetic blueprint of the virus is RNA instead of DNA so it gives infected host cells instructions to read the template and translate it to replicate the virus. Each infected cell may produce and release millions of copies of the virus which can infect other neighboring cells and people when the viral particles are released from the airways (i.e., via coughing or sneezing). |
| - | + | ||
=Spike Protein= | =Spike Protein= | ||
| - | |||
The spike protein[https://en.wikipedia.org/wiki/Coronavirus_spike_protein] is a glycoprotein that lies on the surface of SARS-CoV-2 to facilitate the entry of the virus into host cells. More specifically, once SARS-CoV-2 has entered the respiratory system, its spike protein binds to the ACE2 receptor (Angiotensin Converting Enzyme Receptor 2) in the lungs. The spike protein is cleaved into two functional subunits: S1 and S2. S1 contains the RBDs (receptor binding domains). S2 plays a key role in membrane fusion between the virus and host cell, allowing viral RNA to enter. | The spike protein[https://en.wikipedia.org/wiki/Coronavirus_spike_protein] is a glycoprotein that lies on the surface of SARS-CoV-2 to facilitate the entry of the virus into host cells. More specifically, once SARS-CoV-2 has entered the respiratory system, its spike protein binds to the ACE2 receptor (Angiotensin Converting Enzyme Receptor 2) in the lungs. The spike protein is cleaved into two functional subunits: S1 and S2. S1 contains the RBDs (receptor binding domains). S2 plays a key role in membrane fusion between the virus and host cell, allowing viral RNA to enter. | ||
| Line 20: | Line 14: | ||
===''Receptor Binding Domains''=== | ===''Receptor Binding Domains''=== | ||
The spike protein is a homotrimer with three Receptor Binding Domains <scene name='10/1076042/Overallspikelabeledrbds/2'>(RBDs)</scene>, containing the binding site. The spike protein’s RBD is the region where it attaches to the human ACE2 receptor. A <scene name='10/1076041/Spike_protein_only/3'>closer look</scene> at the RBDs reveal a mostly beta-sheet secondary structure with a handful of helices scattered about. There are also a couple variable loops where a number of interactions take place as well. These interactions are specific to the binding ligand and will be talked about in depth later. | The spike protein is a homotrimer with three Receptor Binding Domains <scene name='10/1076042/Overallspikelabeledrbds/2'>(RBDs)</scene>, containing the binding site. The spike protein’s RBD is the region where it attaches to the human ACE2 receptor. A <scene name='10/1076041/Spike_protein_only/3'>closer look</scene> at the RBDs reveal a mostly beta-sheet secondary structure with a handful of helices scattered about. There are also a couple variable loops where a number of interactions take place as well. These interactions are specific to the binding ligand and will be talked about in depth later. | ||
| - | |||
| - | |||
=ACE2 Receptor= | =ACE2 Receptor= | ||
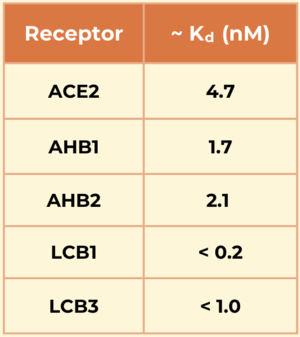
<scene name='10/1076041/Ace2_only/4'>ACE2,</scene> the natural receptor for the spike protein, is a membrane-bound protein located in the lungs and made up of 805 amino acids. Its <scene name='10/1076041/Ace2_interactions/5'>binding site</scene> is located within a single extracellular N-terminal domain. Residues 26-43 in this region form an alpha helix which makes the majority of interactions with the RBD of the spike protein. These interactions are stabilized by hydrogen bonding[https://en.wikipedia.org/wiki/Hydrogen_bond] and van der Waals forces[https://en.wikipedia.org/wiki/Van_der_Waals_force]. | <scene name='10/1076041/Ace2_only/4'>ACE2,</scene> the natural receptor for the spike protein, is a membrane-bound protein located in the lungs and made up of 805 amino acids. Its <scene name='10/1076041/Ace2_interactions/5'>binding site</scene> is located within a single extracellular N-terminal domain. Residues 26-43 in this region form an alpha helix which makes the majority of interactions with the RBD of the spike protein. These interactions are stabilized by hydrogen bonding[https://en.wikipedia.org/wiki/Hydrogen_bond] and van der Waals forces[https://en.wikipedia.org/wiki/Van_der_Waals_force]. | ||
| - | |||
=Mini Protein Inhibitors= | =Mini Protein Inhibitors= | ||
| - | |||
===AHB2=== | ===AHB2=== | ||
<scene name='10/1076041/Ahb2_only/4'>AHB2</scene> is a stable miniprotein inhibitor that competes with the ACE2 receptor. It was developed using the Rosetta blueprint builder that incorporates the alpha helix in ACE2 (residues 23-46) into small, stable protein scaffolds. This molecule utilizes van der Waals interactions in its <scene name='10/1076041/Ahb2_stabilizing_vdw/3'>hydrophobic core</scene> to stabilize. <scene name='10/1076041/Ahb2_interactions/3'>AHB2's interactions</scene> show an increased number of interactions between the spike and minibinder. This allows it to compete with the ACE2 receptor more effectively, preventing it from binding with the spike protein.<ref name="Cao">PMID:32907861</ref> | <scene name='10/1076041/Ahb2_only/4'>AHB2</scene> is a stable miniprotein inhibitor that competes with the ACE2 receptor. It was developed using the Rosetta blueprint builder that incorporates the alpha helix in ACE2 (residues 23-46) into small, stable protein scaffolds. This molecule utilizes van der Waals interactions in its <scene name='10/1076041/Ahb2_stabilizing_vdw/3'>hydrophobic core</scene> to stabilize. <scene name='10/1076041/Ahb2_interactions/3'>AHB2's interactions</scene> show an increased number of interactions between the spike and minibinder. This allows it to compete with the ACE2 receptor more effectively, preventing it from binding with the spike protein.<ref name="Cao">PMID:32907861</ref> | ||
| - | |||
| - | |||
===LCBs=== | ===LCBs=== | ||
<scene name='10/1076041/Overall_lcb1_spike/2'>LCB1</scene> is a fully de novo three-helix mini protein that acts as a competitive inhibitor, competing with ACE2 to bind to the spike protein. Unlike ABH2, a de novo design approach was taken to design LCB1, making it more compact with optimal interactions with the RBD. Its <scene name='10/1076041/Lcb1_interactions/5'>binding interactions</scene> show a large number of hydrogen bonds as well as a small hydrophobic effect promoting binding even more so. | <scene name='10/1076041/Overall_lcb1_spike/2'>LCB1</scene> is a fully de novo three-helix mini protein that acts as a competitive inhibitor, competing with ACE2 to bind to the spike protein. Unlike ABH2, a de novo design approach was taken to design LCB1, making it more compact with optimal interactions with the RBD. Its <scene name='10/1076041/Lcb1_interactions/5'>binding interactions</scene> show a large number of hydrogen bonds as well as a small hydrophobic effect promoting binding even more so. | ||
| - | |||
Another mini protein inhibitor, <scene name='10/1076041/Lcb3_only/3'>LCB3,</scene> was developed using the same computational method as LCB1 and binds to the same region of the RBD, except in the opposite direction. Its <scene name='10/1076041/Lcb3xspikecorlabel_intxn/1'>binding interactions</scene> also show a large number of hydrogen bonds. However, LCB1 has more surface area and fits more precisely with its computational model, giving it a slightly higher binding affinity. | Another mini protein inhibitor, <scene name='10/1076041/Lcb3_only/3'>LCB3,</scene> was developed using the same computational method as LCB1 and binds to the same region of the RBD, except in the opposite direction. Its <scene name='10/1076041/Lcb3xspikecorlabel_intxn/1'>binding interactions</scene> also show a large number of hydrogen bonds. However, LCB1 has more surface area and fits more precisely with its computational model, giving it a slightly higher binding affinity. | ||
Revision as of 14:53, 26 April 2025
De novo miniprotein Covid therapeutic
| |||||||||||