We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Carson Powers/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
=Introduction= | =Introduction= | ||
| - | SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA virus that is responsible for the highly infectious respiratory disease, COVID-19. It first emerged late in 2019 and quickly spread around the world, earning the rank of "pandemic" by March of the following year. Given the severity of the situation the entire world was in, researchers around the globe began researching the virus and methods to prevent its spread. One such method looked to stop the initial infection by preventing the endocytosis of the virus itself through direct competition (for the virus) with the receptor to which it binds. Given this is a respiratory infection, the mode by which to give this competitive inhibitor was designed to be a nose-spray. Initial research looked into the use of antibodies to accomplish this task but found antibodies were not stable enough for this mode. But the use of a new tool changed the game. Using a protein-predicting software, generated proteins solved the stability issue and greatly increased binding affinity. Not only has this tool led to a major step forward in the treatment of COVID-19, but it can even be applied to other infections, marking a new era in the development of therapeutics. | + | SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) is an RNA virus that is responsible for the highly infectious respiratory disease, COVID-19. It first emerged late in 2019 and quickly spread around the world, earning the rank of "pandemic" by March of the following year. Given the severity of the situation the entire world was in, researchers around the globe began researching the virus and methods to prevent its spread. One such method looked to stop the initial infection by preventing the endocytosis of the virus itself through direct competition (for the virus) with the receptor to which it binds. Given this is a respiratory infection, the mode by which to give this competitive inhibitor was designed to be a nose-spray. Initial research looked into the use of antibodies to accomplish this task but found antibodies were not stable enough for this mode. But the use of a new tool changed the game. Using a protein-predicting software, generated proteins, known as miniproteins or mini binders due to their relatively small size, solved the stability issue and greatly increased binding affinity. Not only has this tool led to a major step forward in the treatment of COVID-19, but it can even be applied to other infections, marking a new era in the development of therapeutics. |
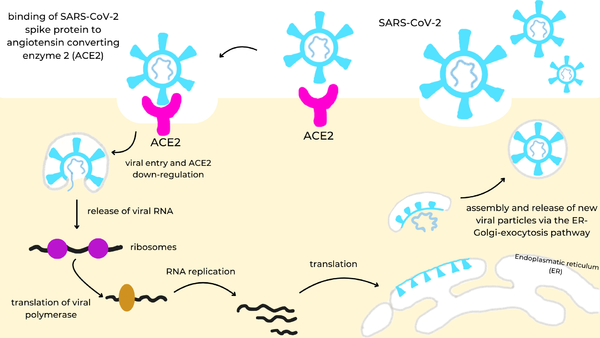
[[Image:spike_infection_cycle.png|600px|right|SARS-CoV-2 infection cycle: the viral spike protein (cyan) binds to the ACE2 receptor (magenta) on the cell surface, triggering endocytosis and ACE2 down-regulation, release of viral RNA into the cytosol, translation of the viral polymerase and genome replication, synthesis of structural proteins in the endoplasmic reticulum (ER), and assembly and release of new virions via the ER-Golgi-exocytosis pathway.]] | [[Image:spike_infection_cycle.png|600px|right|SARS-CoV-2 infection cycle: the viral spike protein (cyan) binds to the ACE2 receptor (magenta) on the cell surface, triggering endocytosis and ACE2 down-regulation, release of viral RNA into the cytosol, translation of the viral polymerase and genome replication, synthesis of structural proteins in the endoplasmic reticulum (ER), and assembly and release of new virions via the ER-Golgi-exocytosis pathway.]] | ||
| Line 19: | Line 19: | ||
=Mini Protein Inhibitors= | =Mini Protein Inhibitors= | ||
===AHB2=== | ===AHB2=== | ||
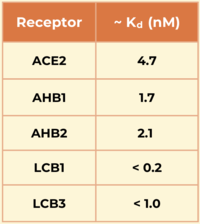
| - | <scene name='10/1076041/Ahb2_only/4'>AHB2</scene> is a | + | <scene name='10/1076041/Ahb2_only/4'>AHB2</scene> is a miniprotein inhibitor that competes with the ACE2 receptor. It was developed using the Rosetta blueprint builder that incorporates the alpha helix in ACE2 (residues 23-46) into small, stable protein scaffolds. This molecule utilizes Van der Waals interactions in its <scene name='10/1076041/Ahb2_stabilizing_vdw/3'>hydrophobic core</scene> to stabilize. <scene name='10/1076041/Ahb2_interactions/3'>AHB2's interactions</scene> show an increased number of interactions between the spike and minibinder. This allows it to compete with the ACE2 receptor more effectively, preventing it from binding with the spike protein.<ref name="Cao">PMID:32907861</ref> |
===LCBs=== | ===LCBs=== | ||
Revision as of 21:35, 27 April 2025
De Novo Miniprotein COVID-19 Therapeutic
| |||||||||||