We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Marcos Ngo/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='7rds' size='340' side='right'caption='[[7rds]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='7rds' size='340' side='right'caption='[[7rds]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Function and Background== | == Function and Background== | ||
| - | DNA glycosylases search the entire genome for DNA lesions. These highly selective enzymes recognize a damaged base and remove it. There are four super families of glycosylases: Udg, Nth, Nei, and AGG. '''hNTHL1''' or '''human Endonuclease III''' (Nth family) is a 34 kDa bifunctional DNA glycosylase involved in the base excision repair (BER) process. A bifunctional glycosylase refers to the ability to '''excise''' damaged bases and '''cleave''' the backbone. hNTHL1 preferentially excises oxidized pyrimidines. Thymine glycol (Tg) is the preferred substrate. Upon encountering this damaged base, the protein severs the N-glycosidic bond, | + | DNA glycosylases search the entire genome for DNA lesions. These highly selective enzymes recognize a damaged base and remove it. There are four super families of glycosylases: Udg, Nth, Nei, and AGG. '''hNTHL1''' or '''human Endonuclease III''' (Nth family) is a 34 kDa bifunctional DNA glycosylase belonging to the HhH (Helix-Hairpin-Helix) superfamily. |
| + | It is involved in the base excision repair (BER) process. A bifunctional glycosylase refers to the ability to '''excise''' damaged bases and '''cleave''' the backbone. hNTHL1 preferentially excises oxidized pyrimidines. Thymine glycol (Tg) is the preferred substrate. Upon encountering this damaged base, the protein severs the N-glycosidic bond, leaving an apurinic site. From here, the backbone is cleaved via beta elimination, which leaves a 3’ aldehyde and creates a single-strand break. Next, the DNA is handed off to Apurinic endonuclease 1 or polynucleotide kinase, leaving a free 3′ hydroxyl for DNA polymerase β to insert the correct nucleotide. Finally, the nick is sealed by the DNA ligase IIIα <ref>PMID:34871433</ref><ref>PMID:20005182</ref><ref>PMID:9295348</ref>. | ||
The gene encoding hNTHL1 is NTHL1, which is located on chromosome 16. It is widely expressed across tissues, with the highest levels observed in the heart. This elevated expression may reflect the heart’s high demand for ATP production, which generates significant oxidative stress and thus increases reliance on base excision repair (BER) proteins for genome maintenance. Additionally, hNTHL1 expression is regulated during the cell cycle, with transcription levels rising during the early and mid S phases <ref>https://www.uniprot.org/uniprotkb/P78549/entry</ref><ref>PMID:8990169</ref><ref>PMID:10882850</ref>. | The gene encoding hNTHL1 is NTHL1, which is located on chromosome 16. It is widely expressed across tissues, with the highest levels observed in the heart. This elevated expression may reflect the heart’s high demand for ATP production, which generates significant oxidative stress and thus increases reliance on base excision repair (BER) proteins for genome maintenance. Additionally, hNTHL1 expression is regulated during the cell cycle, with transcription levels rising during the early and mid S phases <ref>https://www.uniprot.org/uniprotkb/P78549/entry</ref><ref>PMID:8990169</ref><ref>PMID:10882850</ref>. | ||
| Line 12: | Line 13: | ||
DNA glycosylases remove damaged bases through a “pinch, push, plug, and pull” mechanism. First, the DNA is “pinched” by the enzyme, which destabilizes the helix. Next, they use a wedge amino acid to “push” the lesion out of the helix. While the lesion is being flipped out, another amino acid “plugs” into the helix to fill the gap and maintain the structure of the helix. Finally, the lesion is “pulled” into the active site to allow for lesion removal <ref>https://scholarworks.uvm.edu/cgi/viewcontent.cgi?article=2160&context=graddis</ref><ref>PMID:20469926</ref><ref>PMID:12220189</ref>. | DNA glycosylases remove damaged bases through a “pinch, push, plug, and pull” mechanism. First, the DNA is “pinched” by the enzyme, which destabilizes the helix. Next, they use a wedge amino acid to “push” the lesion out of the helix. While the lesion is being flipped out, another amino acid “plugs” into the helix to fill the gap and maintain the structure of the helix. Finally, the lesion is “pulled” into the active site to allow for lesion removal <ref>https://scholarworks.uvm.edu/cgi/viewcontent.cgi?article=2160&context=graddis</ref><ref>PMID:20469926</ref><ref>PMID:12220189</ref>. | ||
| - | Recent studies | + | Recent studies examined how hNTHL1 initiates BER of oxidative lesions in nucleosomal DNA. When lesions are positioned away from the histone core, hNTHL1 processes them with nearly the same efficiency as in naked DNA (without histones). When lesions faced inward toward the histone core, repair was initially poor but improved significantly at higher hNTHL1 concentrations. Additionally, sections near the edge of the nucleosome were repaired more efficiently than those near the nucleosome center. This suggests that both the partial unwrapping of DNA from the histone core and the positioning of the lesion outward from the nucleosome edge allow hNTHL1 to efficiently access and repair DNA lesions <ref>PMID:17923696</ref>. |
== Structural Highlights == | == Structural Highlights == | ||
| Line 21: | Line 22: | ||
</table> | </table> | ||
| - | + | hNTHL1 consists of <scene name='10/1077482/Two_domains/2'>two alpha-helical domains</scene> connected by two linkers. <scene name='10/1077482/Ncfedomain1/5'>Domain 1</scene> has the iron sulfur cluster, N- and C-termini, and a catalytic residue ('''Asp 239'''). <scene name='10/1077482/Domain2features/3'>Domain 2</scene> has six helical barrels, hairpin-helix-hairpin, and the final catalytic residue ('''Lys 220'''). The <scene name='10/1077482/Proglyhhh/1'>HhH</scene> motif has a characteristic glycine and proline-rich loop. The HhH allows for hydrogen bond interactions with the DNA backbone <ref>PMID:12840008</ref><ref>https://scholarworks.uvm.edu/cgi/viewcontent.cgi?article=2160&context=graddis</ref><ref>PMID:1283262</ref>. | |
This structure is captured in an <scene name='10/1077482/Open_conformation/1'>open conformation</scene> where the catalytic residues Lys220 and Asp239 are positioned approximately 25 Å apart, which is too far for catalysis. This implies that a conformational change is required to assemble the active site. To find the closed conformation, an engineered chimera was made by swapping the <scene name='10/1077482/Linker1/1'>flexible interdomain linker</scene> in human NTHL1 with a shorter, more rigid linker from a bacterial homolog. The <scene name='10/1077482/Chimera/1'>hNTHL1Δ63 chimera</scene> structure adopts a closed conformation where Lys220 and Asp239 are approximately 5 Å apart, which mimics the configuration seen in catalytically active homologs. The linker is not fully modeled due to disorder in the electron density map <ref>PMID:34871433</ref>. | This structure is captured in an <scene name='10/1077482/Open_conformation/1'>open conformation</scene> where the catalytic residues Lys220 and Asp239 are positioned approximately 25 Å apart, which is too far for catalysis. This implies that a conformational change is required to assemble the active site. To find the closed conformation, an engineered chimera was made by swapping the <scene name='10/1077482/Linker1/1'>flexible interdomain linker</scene> in human NTHL1 with a shorter, more rigid linker from a bacterial homolog. The <scene name='10/1077482/Chimera/1'>hNTHL1Δ63 chimera</scene> structure adopts a closed conformation where Lys220 and Asp239 are approximately 5 Å apart, which mimics the configuration seen in catalytically active homologs. The linker is not fully modeled due to disorder in the electron density map <ref>PMID:34871433</ref>. | ||
| Line 29: | Line 30: | ||
The function of the <scene name='10/1077482/N-terminus_AlphaFold/3'>N-terminus</scene> (AlphaFold Prediction) of hNTHL1 has been a subject of study <ref>https://www.nature.com/articles/s41586-021-03819-2/</ref><ref>PMID:37933859</ref>. It is theorized that the N-terminus, which is extended compared to homologs, functions as a means to remain bound to DNA, protecting the labile abasic site while waiting for its handoff with APE1. This was found through truncation of the N-terminal region (residues 1-96), which revealed that deletion of 55, 75, or 80 residues from the N-terminus resulted in a four to fivefold increase in catalytic activity. The rate-limiting step in hNTHL1's reaction is the release of the free 3’ aldehyde <ref>PMID:12144783</ref>. | The function of the <scene name='10/1077482/N-terminus_AlphaFold/3'>N-terminus</scene> (AlphaFold Prediction) of hNTHL1 has been a subject of study <ref>https://www.nature.com/articles/s41586-021-03819-2/</ref><ref>PMID:37933859</ref>. It is theorized that the N-terminus, which is extended compared to homologs, functions as a means to remain bound to DNA, protecting the labile abasic site while waiting for its handoff with APE1. This was found through truncation of the N-terminal region (residues 1-96), which revealed that deletion of 55, 75, or 80 residues from the N-terminus resulted in a four to fivefold increase in catalytic activity. The rate-limiting step in hNTHL1's reaction is the release of the free 3’ aldehyde <ref>PMID:12144783</ref>. | ||
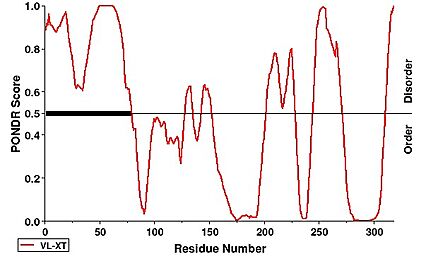
| - | Notably, the first 63 residues of | + | Notably, the first 63 residues of the N-terminus were not modeled due to disorder. This can be observed under a PONDR prediction |
[[Image:PONDR.jpg|438 × 271px|'''PONDR Disorder Prediction''']] <ref>https://www.pondr.com/</ref>. | [[Image:PONDR.jpg|438 × 271px|'''PONDR Disorder Prediction''']] <ref>https://www.pondr.com/</ref>. | ||
Revision as of 00:29, 28 April 2025
Human NTHL1
| |||||||||||