We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
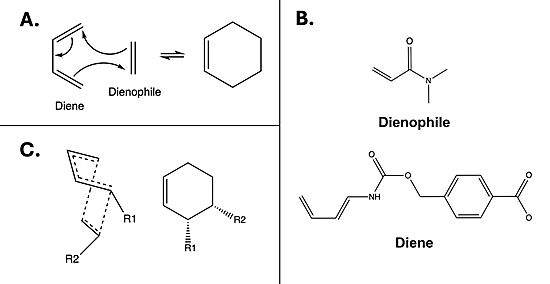
| - | The Diels Alderase aims to catalyze the Diels-Alder reaction between 4-carboxybenzyl-trans-1,3-butadiene-1-carbamate and N,N-dimethylacrylamide for use in synthetic organic chemistry. Specifically, the enzyme surpasses uncatalyzed reactions by generating a product that is entirely [https://en.wikipedia.org/wiki/Stereoselectivity#:~:text=In%20chemistry%2C%20stereoselectivity%20is%20the,of%20a%20pre%2Dexisting%20one. stereoselective] for the 3R,4S endo form. The Diels-Alderase was built using ''de novo'' enzyme design, using computational modeling and refinement through collaborative problem-solving from online users. The first generation Diels-Alderase was made using the [https://en.wikipedia.org/wiki/Rosetta@home Rosetta] computational design program, where a potential active site was built and tested against a library of scaffold proteins. Later, as the active site was perfected, future generations of the Diels-Alderase were made using an online protein folding game called [https://en.wikipedia.org/wiki/Foldit Foldit,] where players competed to improve binding efficiency by completing various challenges.<ref name="Eiben"/> | + | The Diels-Alderase aims to catalyze the Diels-Alder reaction [Fig. 1A] between 4-carboxybenzyl-trans-1,3-butadiene-1-carbamate and N,N-dimethylacrylamide [Fig. 1B] for use in synthetic organic chemistry. Specifically, the enzyme surpasses uncatalyzed reactions by generating a product that is entirely [https://en.wikipedia.org/wiki/Stereoselectivity#:~:text=In%20chemistry%2C%20stereoselectivity%20is%20the,of%20a%20pre%2Dexisting%20one. stereoselective] for the 3R,4S endo form [Fig.1C]. The Diels-Alderase was built using ''de novo'' enzyme design, using computational modeling and refinement through collaborative problem-solving from online users. The first generation Diels-Alderase was made using the [https://en.wikipedia.org/wiki/Rosetta@home Rosetta] computational design program, where a potential active site was built and tested against a library of scaffold proteins. Later, as the active site was perfected, future generations of the Diels-Alderase were made using an online protein folding game called [https://en.wikipedia.org/wiki/Foldit Foldit,] where players competed to improve binding efficiency by completing various challenges.<ref name="Eiben"/> |
| + | |||
| + | [[Image:DielsAlderasecomboinfo Large.jpeg|550px|left|thumb|Figure 1. A) Example mechanism of a simple Diels-Alder reaction. B) Diels-Alderase substrates. Diene is 4-carboxybenzyl trans-1,3-butadiene-1-carbamate; dienophile is N,N- dimethylacrylamide. C) Illustration of 3R, 4S endo stereoisomerism, which the Diels-Alderase is selective for.]] | ||
| - | [[Image:Dielsalderasesubstrates.jpeg|350px|left|thumb|Figure 1. Diels-Alderase substrates. Diene is 4-carboxybenzyl trans-1,3-butadiene-1-carbamate; dienophile is N,N- dimethylacrylamide.]] | ||
The Diels-Alderase was designed to connect a diene and dienophile in a [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction Diels-Alder reaction.] It accomplishes this by decreasing the energy gap between the dienophile’s lowest unoccupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (LUMO)] and the diene’s highest occupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (HOMO)] in the transition state.<ref name="Siegel">PMID:20647463</ref> The current most active form of the Diels-Alderase is modelled under the PDB code [https://www.rcsb.org/structure/4O5T 4o5t]. | The Diels-Alderase was designed to connect a diene and dienophile in a [https://en.wikipedia.org/wiki/Diels%E2%80%93Alder_reaction Diels-Alder reaction.] It accomplishes this by decreasing the energy gap between the dienophile’s lowest unoccupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (LUMO)] and the diene’s highest occupied molecular orbital [https://en.wikipedia.org/wiki/HOMO_and_LUMO (HOMO)] in the transition state.<ref name="Siegel">PMID:20647463</ref> The current most active form of the Diels-Alderase is modelled under the PDB code [https://www.rcsb.org/structure/4O5T 4o5t]. | ||
| - | [[Image:DAstereoselectivity.png|350px|left|thumb|Figure.x]] | ||
| - | [[Image:SampleDAmech.jpeg|350px|right|thumb|Figure X.]] | ||
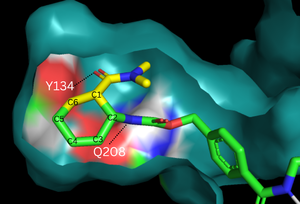
| - | The binding pocket of 4o5t is selective for two substrates, 4-carboxybenzyl trans-1,3-butadiene-1-carbamate ([https://en.wikipedia.org/wiki/Diene diene]) and N,N- dimethylacrylamide (dienophile). These substrates are shown as a single, combined | + | The binding pocket of 4o5t is selective for two substrates, 4-carboxybenzyl trans-1,3-butadiene-1-carbamate ([https://en.wikipedia.org/wiki/Diene diene]) and N,N- dimethylacrylamide (dienophile). These substrates are shown as a single, combined ligand, 4-{[2-(phosphonooxy)ethyl]carbamoyl}benzyl [(1R,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamate, in the protein model. The binding site contains a [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] donor (Tyr134) which lowers the LUMO energy and stabilizes the negative charge on the dienophile and a hydrogen bond acceptor (Glu208) that increases the HOMO energy and stabilizes the positive charge on the diene.<ref name="Siegel"/> Both of these H-bonding interactions work to stabilize the transition state, while also orienting the substrates in optimal conformations for reacting. |
Overall, the Diels-Alderase stimulates improvement in synthetic laboratories and demonstrates early success in the now-prominent world of [https://www.nobelprize.org/prizes/chemistry/2024/press-release/ computational enzyme design.] | Overall, the Diels-Alderase stimulates improvement in synthetic laboratories and demonstrates early success in the now-prominent world of [https://www.nobelprize.org/prizes/chemistry/2024/press-release/ computational enzyme design.] | ||
| Line 23: | Line 22: | ||
====Helix Cap==== | ====Helix Cap==== | ||
In the evolution process, a 16-residue [https://proteopedia.org/wiki/index.php/Alpha_helix alpha-helix] <scene name='10/1075254/Alpha_helix_highlighted/1'>cap</scene> to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction,” decreasing the ''K<sub>m</sub>'' of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.<ref name="Eiben">PMID:22267011</ref> | In the evolution process, a 16-residue [https://proteopedia.org/wiki/index.php/Alpha_helix alpha-helix] <scene name='10/1075254/Alpha_helix_highlighted/1'>cap</scene> to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction,” decreasing the ''K<sub>m</sub>'' of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.<ref name="Eiben">PMID:22267011</ref> | ||
| - | |||
| - | == Mechanism == | ||
| - | [[Image:Resizedmechanism.png|500px|left|thumb|Figure 3. Active site mechanism]] | ||
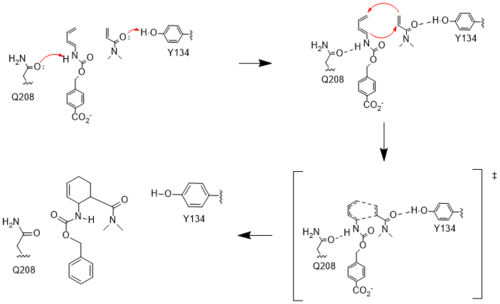
| - | The key to this enzyme’s success as a catalyst lies in its ability to lower the energy gap between reactants. To accomplish this, the two active site residues, Tyr134 and Glu208, use hydrogen bonding to assist the reaction in a variety of ways. First, it allows specific binding of the ligand in the active site, selecting for molecules with certain stereochemistry at and around the catalytic residues, specifically the carbamate and carbonyl of the diene and dienophile, respectively. This promotes the reaction by stabilizing the molecules in close proximity to one another, which also promotes the characteristic stereoselectivity.<ref name="Siegel"/> Second, the bonds affect the energetics of the molecules. By donating a hydrogen to the carbonyl of dienophile, Tyr134 helps to decrease the electron density around the molecule, lowering the energy of the lowest unoccupied molecular orbital (LUMO). Conversely, by abstracting the hydrogen from the carbamate of the dienophile, Glu208 increases the electron density and thus the energy of the highest occupied molecular orbital (HOMO). By closing the gap between these orbitals, the enzyme lowers the activation energy required to allow the orbitals to react. Finally, these interactions help to stabilize the accumulated charges in the transition state. By decreasing electron density in the dienophile, Tyr134 helps to stabilize the accumulated negative charge in the transition state. The Glu208, then, helps stabilize the accumulated positive charge by increasing the electron density of the diene. Calculations predict that this helps to stabilize the transition state by nearly 5 kcal/mol.<ref name="Siegel"/> All together, these interactions make it much easier for the reaction to proceed in a very stereoselective and favorable manner. | ||
| + | == Mechanism == | ||
| + | [[Image:Resizedmechanism.png|500px|left|thumb|Figure 3. Active site mechanism]] | ||
| + | The key to this enzyme’s success as a catalyst lies in its ability to lower the energy gap between reactants. To accomplish this, the two active site residues, Tyr134 and Glu208, use hydrogen bonding to assist the reaction in a variety of ways. First, it allows specific binding of the ligand in the active site, selecting for molecules with certain stereochemistry at and around the catalytic residues, specifically the carbamate and carbonyl of the diene and dienophile, respectively. This promotes the reaction by stabilizing the molecules in close proximity to one another, which also promotes the characteristic stereoselectivity.<ref name="Siegel"/> Second, the bonds affect the energetics of the molecules. By donating a hydrogen to the carbonyl of dienophile, Tyr134 helps to decrease the electron density around the molecule, lowering the energy of the lowest unoccupied molecular orbital (LUMO). Conversely, by abstracting the hydrogen from the carbamate of the dienophile, Glu208 increases the electron density and thus the energy of the highest occupied molecular orbital (HOMO). By closing the gap between these orbitals, the enzyme lowers the activation energy required to allow the orbitals to react. Finally, these interactions help to stabilize the accumulated charges in the transition state. By decreasing electron density in the dienophile, Tyr134 helps to stabilize the accumulated negative charge in the transition state. The Glu208, then, helps stabilize the accumulated positive charge by increasing the electron density of the diene. Calculations predict that this helps to stabilize the transition state by nearly 5 kcal/mol.<ref name="Siegel"/> All together, these interactions make it much easier for the reaction to proceed in a very stereoselective and favorable manner. | ||
Revision as of 01:59, 28 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Diels-Alderase
| |||||||||||