Sandbox2O14
From Proteopedia
| Line 47: | Line 47: | ||
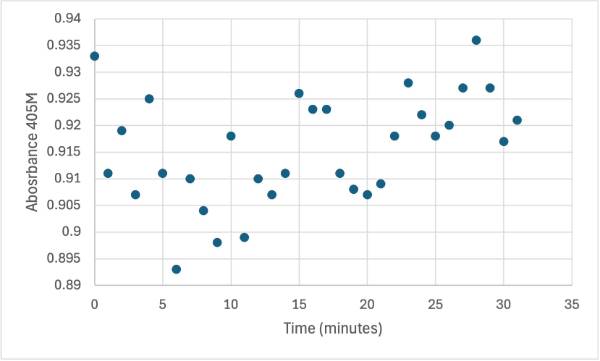
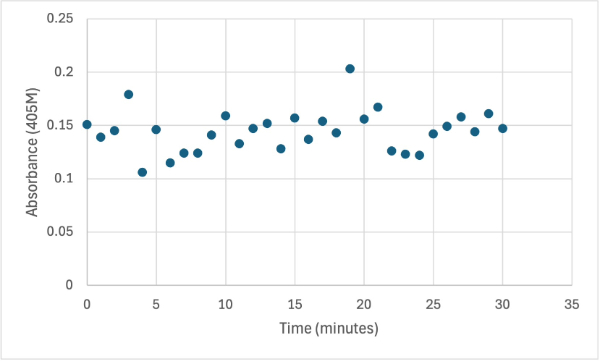
== Figure 4 == | == Figure 4 == | ||
[[Image:1_30mg1stSMALL.jpg]] | [[Image:1_30mg1stSMALL.jpg]] | ||
| + | |||
Protein test using 10µL of protein from elution and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | ||
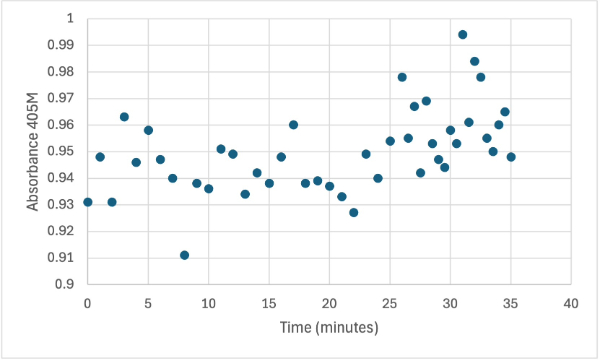
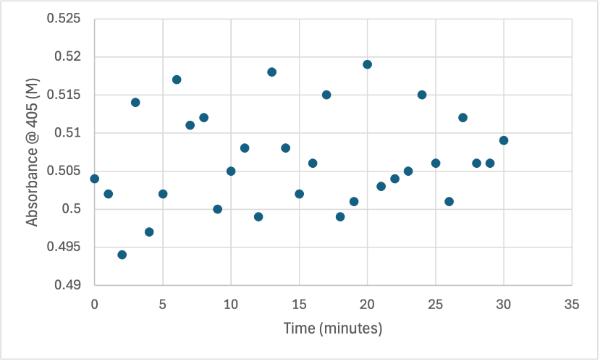
== Figure 5 == | == Figure 5 == | ||
| - | [[Image: | + | [[Image:2_30mg2ndelutSMALL.jpg]] |
| + | |||
Protein test using 10µL of protein from elution 2 and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 2 and 30mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | ||
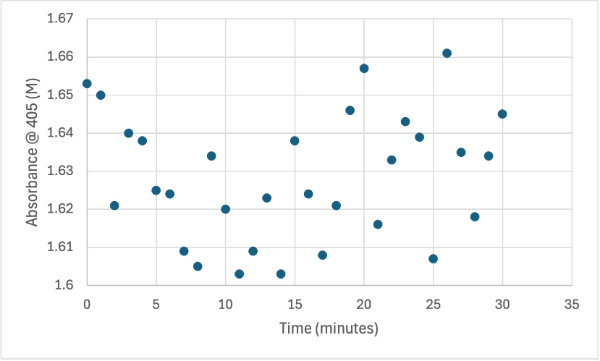
== Figure 6 == | == Figure 6 == | ||
| - | [[Image: | + | [[Image:3_ph560SMALL.jpg]] |
| + | |||
Protein test using 10µL of protein from elution 1 and 30mg of PNPP and 3mL of PNPP solution, changing the pH to about 5.60. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 1 and 30mg of PNPP and 3mL of PNPP solution, changing the pH to about 5.60. Measuring absorbance at 405M. | ||
| Line 63: | Line 66: | ||
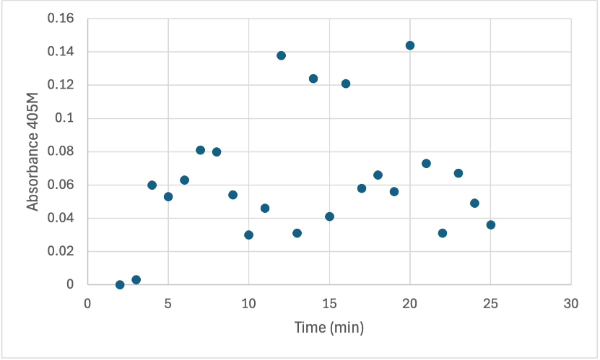
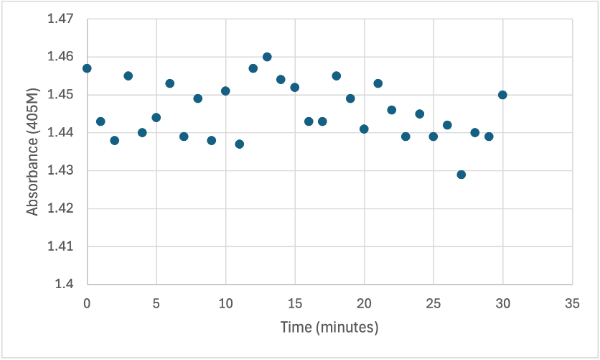
== Figure 8 == | == Figure 8 == | ||
| - | [[Image: | + | [[Image:4_3mgElut1SMALL.jpg]] |
| + | |||
Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution. Measuring absorbance at 405M. | ||
== Figure 9 == | == Figure 9 == | ||
| - | [[Image: | + | [[Image:5_3mg1HClSMALL.jpg]] |
| + | |||
Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 1 and 3mg of PNPP and 3mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M. | ||
== Figure 10 == | == Figure 10 == | ||
| - | [[Image: | + | [[Image:6_10mg10mLPNPASMALL.png]] |
| + | |||
Protein test using 10µL of protein from elution 1 and 10mg of PNPA and 10mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 1 and 10mg of PNPA and 10mL of PNPP solution and 1 drop of HCL with a pH of about 10.50. Measuring absorbance at 405M. | ||
| Line 77: | Line 83: | ||
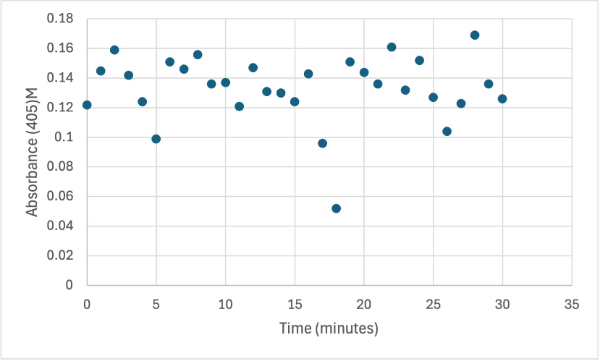
== Figure 11 == | == Figure 11 == | ||
| - | [[Image: | + | [[Image:7_15mgPNPPSMALL.png]] |
| + | |||
Protein test using 10µL of protein from elution 1 and 15mg of PNPA and 3mL of PNPP solution. Measuring absorbance at 405M. | Protein test using 10µL of protein from elution 1 and 15mg of PNPA and 3mL of PNPP solution. Measuring absorbance at 405M. | ||
== Figure 12 == | == Figure 12 == | ||
| - | [[Image: | + | [[Image:8_PNPAaddproteinSMALL.jpg]] |
| + | |||
Protein test with increasing protein solution of 5µL every minute from elution 1 with 15mg of PNPA and 3mL PNPP solution. The pH of the solution is 8.35 and it was measuring absorbance at 405M. | Protein test with increasing protein solution of 5µL every minute from elution 1 with 15mg of PNPA and 3mL PNPP solution. The pH of the solution is 8.35 and it was measuring absorbance at 405M. | ||
Revision as of 02:42, 28 April 2025
2O14 Structure and Proposed Functionality
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this.)
2O14 is a monomeric protein complex that originates from the bacterial species Bacillus subtilis and has a mass of 41.79 kDa. Using InterPro this protein was assumed to be like Rhamnogalacturan acetylesterase, which falls under the SGNH Hydrolase Superfamily with structural and sequential similarities to lipases, esterases, along with similar functional domains to galactose-binding. With the research performed with current protein databases available, the evidence suggests that this protein removes acetyl groups from the backbone proteins by hydrolyzing the acetyl esters. It accomplishes this most likely through the catalytic triad of serine-histidine-aspartic acid.
| |||||||||||
Contents |
Figure 1
Lowest Calculated affinity image from SwissDock of -6.133 kcal/mol, with the ligand 2-acetamido-2-deoxy-beta-D-glucopyranose.
Figure 2
The second lowest calculated binding affinity value, -5.629 kcal/mol, predicted bonding site. Believed that the protein’s binding site is in the center between the beta sheet group on top and the alpha helices on the bottom with the ligand 2-acetamido-2-deoxy-beta-D-glucopyranose.
Figure 3
Images shows another potential bonding site on the outside of the protein with a binding affinity of -4.429 kcal/mole with the ligand 2-acetamido-2-deoxy-beta-D-glucopyranose.
References
A) Mølgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan Acetylesterase Elucidates the Structure and Function of a New Family of Hydrolases. Structure 2000, 8 (4), 373–383. https://doi.org/10.1016/S0969-2126(00)00118-0. B) Schoch, C. L.; Ciufo, S.; Domrachev, M.; Hotton, C. L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; Sharma, S.; Soussov, V.; Sullivan, J. P.; Sun, L.; Turner, S.; Karsch-Mizrachi, I. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database 2020, 2020, baaa062. https://doi.org/10.1093/database/baaa062. C) Miesfeld, R. L.; McEvoy, M. M. Biochemistry, 2nd ed.; W. W. Norton & Company, 2021. D) SGNH hydrolase superfamily. InterPro, 2017. https://www.ebi.ac.uk/interpro/entry/InterPro/IPR036514/ E) Bugnon M, Röhrig UF, GOullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024 F) Eberhardt J, Santos-Martins D, Tillack AF, Forli S.. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021