Sandbox323
From Proteopedia
| Line 96: | Line 96: | ||
Absorbance values can be transformed to units of concentration via the Beer-Lambert law. We must accept the approximation of the Molar Extinction Coefficient for PNPA hydrolysis at 428.7nm as 12000 M^-1 cm^-1. An example calculation is supplied in the table. | Absorbance values can be transformed to units of concentration via the Beer-Lambert law. We must accept the approximation of the Molar Extinction Coefficient for PNPA hydrolysis at 428.7nm as 12000 M^-1 cm^-1. An example calculation is supplied in the table. | ||
| - | [[Image:Tablebeerlaw.png| | + | [[Image:Tablebeerlaw.png|400px|left|]] |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 12:54, 28 April 2025
4Q7Q Structure and Proposed Functionality
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this. -Neil Divins)
4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site.
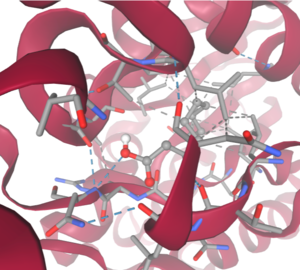
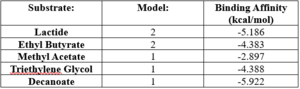
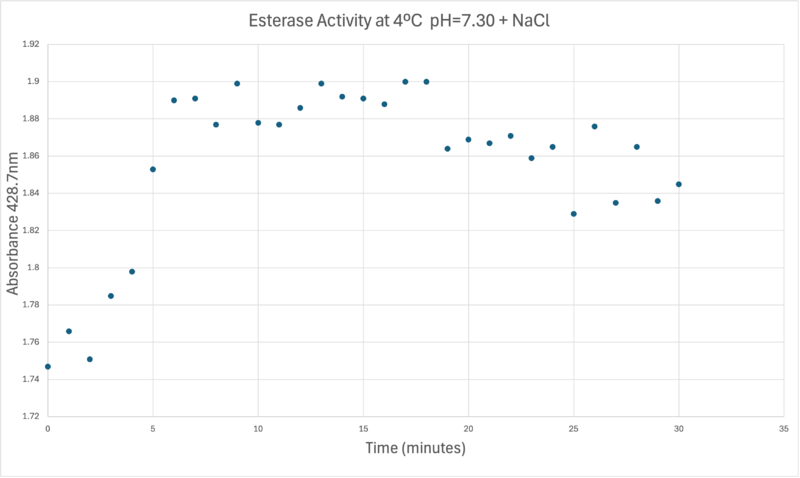
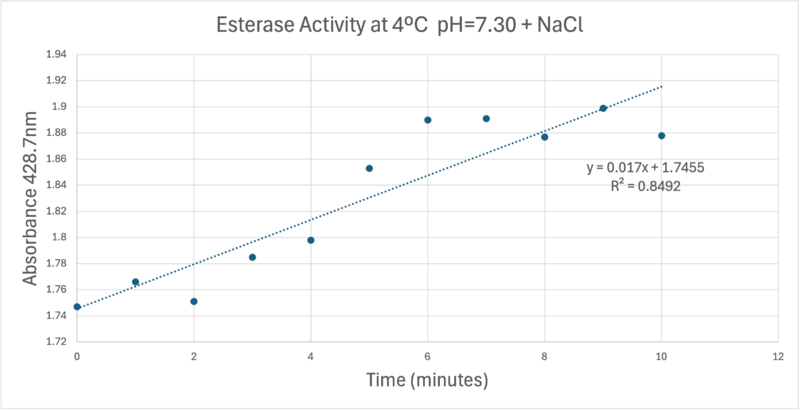
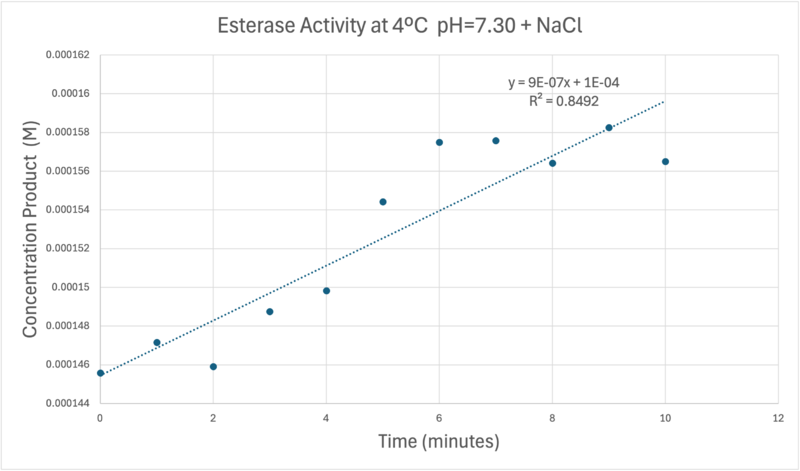
OverviewGeneral Structure and Origins4Q7Q exists as a homodimer Quaternary structure.[1] Analyzing primary and Quaternary structures of 4Q7Q with SPRITE revealed two chains identical in both shape and sequence[2] Each chain is 266 residues long, and the entire complex has a molecular weight of approximately 58.5 kDa.[1].  Primary Sequences of the A and B chains of 4Q7Q.[2]. 4Q7Q proteins primarily appear in bacterial species.[3]. Research reveals how nearly every enzyme with similar sequencing to 4Q7Q is found in bacterium, with a slight exception to eukaryotes.[4] Additionally, the PDB entry for 4Q7Q notes how it potentially can be found in Chitinophaga pinensis, a gram-negative bacterial species which can degrade chitin.[3].[5]. Family and SuperfamilyResearch shows that 4Q7Q is a member of the SGNH Hydrolase protein super family. BLAST and InterPro research suggested 4Q7Q best fits this superfamily, and the known conserved residues seen from SPRITE analysis—Serine, Glycine, Asparagine, and Histidine—line up with those observed throughout this family.[4][6]. Notably, this superfamily is also referred to as the GDSL Hydrolase superfamily.[4][6].  Chimera-generated representation of the A chain of 4Q7Q.[7] Regarding what protein family 4Q7Q belongs to, DALI results suggest it is a part of a sub-family of the greater GDSL/SGNH superfamily. A PDB90% DALI search labels 4Q7Q as a part of the “Lipolytic Protein G-D-S-L Family,” which refers to enzymes that hydrolyze lipid substrates.[8]. Structure/Sequence AnalysisSequence AnalysisThe primary sequence of 4Q7Q shows several conserved sequences between it and esterase-like proteins. A sequence of GDSI—similar to the GDSL sequence seen from its family and superfamily—can be seen between 4Q7Q and enzymes like Isoamyl Acetate-Hydrolyzing Esterase when using DALI.[9]. Other noteworthy conserved sequences between esterases and 4Q7Q include GxND and DGxH.[9]. Conserved sequences of note between 4Q7Q and Esterases. [9]. These enzymes also share similar secondary structures. Segments of alpha-helixes and beta-sheet strands appear and remain nearly entirely conserved throughout esterase analysis. A few conserved coils appear, but these sections do not appear as often as the other two secondary structures. Conserved secondary structures of note between 4Q7Q and Esterases.[9]. Similar conserved sequences could be found between 4Q7Q and lipases. The GDSI, GxND, and DGxH sequences can be seen from lipases like 7BXD.[9] The same secondary structure segments can also be located in the lipases analyzed. Structural AnalysisRight-hand SPRITE analysis revealed 4Q7Q exhibited residues like those seen from enzymes operating with acetyl-like substrates.[2] Specifically, residues Ser. 30, Gly. 69, Asn. 97, Asp. 251, and His. 254 on the A and B chains of 4Q7Q line up with similarly positioned residues on esterases like Platelet-Activating Factor Acetylhyd (PAFA), which exhibited an RMSD of 0.25 angstroms when compared to 4Q7Q.[2] SPRITE-based alignment between motifs from 4Q7Q and PAFA.[2]. Other proteins with similar motifs of note are Thioesterase I and Rhamnogalacturonan Acetylesterase, with RMSD values of 0.46 and 0.61, respectively.[2][6] These alignments focus on the same active site as PAFA did, suggesting the acetyl-like substrates 4Q7Q focuses on are similar to esters. PFAM graphics from DALI revealed significant structural equivalence between 4Q7Q, a lipase-like protein, Rhamnogalacturonan Acetylesterase, and Sialate O-acetylesterase.[9] 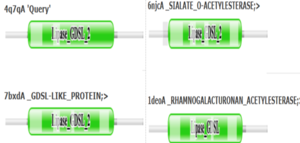 Pfam similarities between 4Q7Q and other enzymes.[9]. 4Q7Q’s superfamily also supports its SPRITE-derived hypothetical functionality. Rhamnogalacturonan Acetylesterase—the enzyme with one of the best SPRITE-based alignment relative to 4Q7Q—is a member of this family.[6][2] Proteins in this family are also known for containing a “unique hydrogen bond network that [stabilizes]” the active site.[2] Proposed FunctionalitySubstrates and Docking AnalysisSwissDock analysis showed a preference for larger molecules, specifically fatty acids. Lactide, Ethyl Butyrate, and Triethylene Glycol exhibited noticeably weak binding affinities to the theorized active site of 4Q7Q. [10][11][12][13] These ligands may be ill-suited to act as substrates for 4Q7Q as they are remarkably polar, and lipids—one of the potential categories of substrates for 4Q7Q—are mostly non-polar.[10][11][12][13] Despite this, these ligands show noticeable hydrophobic interactions with the active site. This implies 4Q7Q uses hydrophobic regions to help guide substrates into the right orientation for enzymatic processes. This also further supports the possibility that 4Q7Q primarily operates with hydrophobic lipid-based substrates. This also explains why Methyl Acetate exhibited a relatively weaker affinity for 4Q7Q, as its smaller structure prevented hydrophobic interactions. Hypothetical FunctionTaking all of the above evidence into consideration, we currently believe 4Q7Q is an enzyme responsible for the hydrolysis of lipid esters and/or fatty acids via a catalytic triad active site. This may suggest 4Q7Q plays an important role in providing energy to the body in a method similar to the beta-oxidation of fatty acids [14] Other studies suggest that the hydrolysis of fatty acids could be involved in fermentation-related processes or even the degradation of aryl lipid esters.[15][5] We also believe 4Q7Q undergoes several significant structural changes during enzymatic activities. Analysis into other members of its family reveal mechanisms wherein serine and histidine residues shift during substrate binding. [5] Specifically, the mechanism of note is similar to the ester hydrolysis or formation of lipases and esterases and is composed of four steps: First, the substrate is bound to the active serine, yielding a tetrahedral intermediate stabilized by the catalytic His and Asp residues. Next, the alcohol is released and an acyl–enzyme complex is formed. Attack of a nucleophile (water in hydrolysis) forms again a tetrahedral intermediate, which after resolution yields the product and free enzyme.[16] The residues involved with this catalytic mechanism are also similar to the residues located in the motif of note from SPRITE analysis.[2][16] Experimental DataSubstrate StructureHydrolysis of SubstrateSpectrophotometry50uL of purified protein from the first elution was added to 3mL of cold 1mg/mL PNPA. Solid sodium chloride was added to achieve a final concentration of 5mM NaCl. The absorbance peak was measured every 60 seconds for 30 minutes. Peak maximum was located at 427.8nm. The sample incubated in an ice bath between measurements. No discernible relationship between time and absorbance was noted. pH of the solution was 10.60. The instrument to measure pH was Vernier Lab Quest 3 with Tris-compatible flat pH sensor calibrated with two points. Theorizing the enzyme would increase activity at the physiological pH of common prokaryotes, we lowered the pH with 2M HCl. 2M NaOH was used to raise the pH due to the weak buffering capacity of the PNPA solution near pH 7. Final pH of the 1mg/mL PNPA solution was 7.30 50uL of purified protein from the first elution was added to 3mL of cold 1mg/mL PNPA at pH=7.30. Chloride ion cofactor was provided by the 2M HCl. A change in absorbance at 428.7nm over time was noted. The time range of the run did not produce a linear response. The absorbance increased for the first ten minutes and plateaued. The time 0 to 10 minutes was used to create the third figure with a more refined linear range. A linear equation of y=9.275x10^-7x+1x10^-4 with a coefficient of determination of 0.8492 was calculated using linear regression. Absorbance values can be transformed to units of concentration via the Beer-Lambert law. We must accept the approximation of the Molar Extinction Coefficient for PNPA hydrolysis at 428.7nm as 12000 M^-1 cm^-1. An example calculation is supplied in the table. References
| ||||||||||||