We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1848
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Function == | == Function == | ||
| - | These minibinders were designed to form stronger connections to the spike protein RBD than ACE2<ref name="Longxing">PMID:32907861</ref>. This section highlights some important residue connections with the RBD that give the minibinders higher affinities for the spike protein than ACE2. | ||
| - | |||
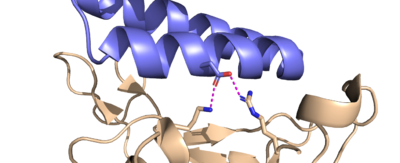
| - | ACE2 binds to the spike protein RBD using these residues. The minibinders are smaller than ACE2, but still form strong connections to the spike protein. This means the minibinders bond to the RBD residues more efficiently than ACE2 giving them a higher affinity for the spike protein <ref name="Longxing">PMID:32907861</ref>. AHB2 was designed to mimic ACE2 while making better use of the RBD residues giving it a higher affinity for the spike protein with an IC50 value of 15.5 nM<ref name="Longxing">PMID:32907861</ref> (AHB2 and RBD binding residues). LCB1 was designed from scratch to bind the most efficiently to the RBD residues giving it the highest affinity with the lowest IC50 value of 23.5 pM<ref name="Longxing">PMID:32907861</ref> (LCB1 and RBD binding residues). LCB3 was designed after LCB1 with the goal of making the minibinder even more efficient, however it ended up having a lower affinity than LCB1 with a higher IC50 value of 40.1 pM<ref name="Longxing">PMID:32907861</ref> (LCB3 and RBD binding residues). | ||
| - | |||
| - | The Glutamine-493 residue on the spike protein is an important residue in showing the differences in strength between ACE2 and the minibinders <ref name="Longxing">PMID:32907861</ref>. ACE2 does not make use of this residue when binding to the RBD. The nearest residues of ACE2, Gln35 and Lys31 do not form any interaction on Gln493 of the spike protein. The AHB2 minibinder, instead forms a Hydrogen bond on the Gln493 residue of the spike protein with its Glu41, increasing affinity to the spike protein. LCB1 forms two hydrogen bonds on Gln493 of the spike protein using two different residues, giving it the highest affinity based on the Gln493 residue. However, these interactions are not required. LCB1 forms no interactions with the Gln493 residue of the RDB previously mentioned, yet it still has a higher affinity to the RBD than AHB2 which forms a hydrogen bond with the Gln493 residue. | ||
| - | |||
| - | Similarly, ACE2 forms a hydrogen bond with the Lys417 from its Asp30 residue. LCB1, however, forms two hydrogen bonds with both the Lys417 and Arg403 of the spike protein from its Asp30 residue. This is a very strong interaction, further contributing to the high affinity of LCB1 to the spike protein. | ||
| + | ==example/sandbox section== | ||
===meow=== | ===meow=== | ||
Revision as of 16:59, 28 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ 4.0 4.1 4.2 Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020 Oct 23;370(6515):426-431. PMID:32907861 doi:10.1126/science.abd9909