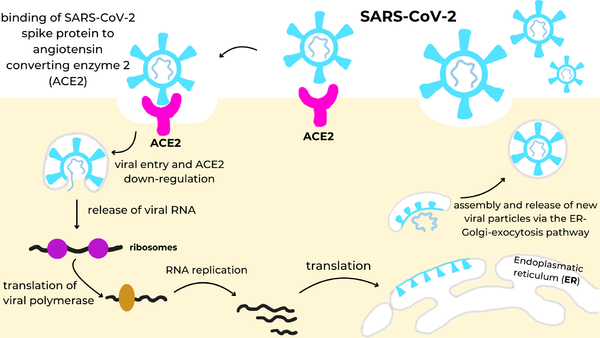
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Carson Powers/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
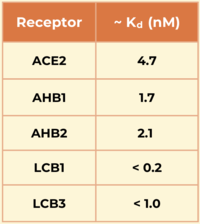
Another miniprotein, <scene name='10/1076041/Lcb3_scene_1/4'>LCB3</scene>, was developed using the same computational method as LCB1 and binds to the same region of the RBD, except in the opposite direction. Its <scene name='10/1076041/Lcb3xspikecorlabel_intxn/1'>binding interactions</scene> also show a large number of hydrogen bonds. However, LCB1 has more surface area and fits more precisely with the spike protein, giving it a slightly higher binding affinity (shown in the table to the right).[[Image:Kd_table.png|200px|right|'''Table 1'''. Dissociation constants (Kd) in nM for SARS-CoV-2 spike RBD binding to ACE2 and engineered minibinders, measured by biolayer interferometry to compare affinities for the spike protein.]] | Another miniprotein, <scene name='10/1076041/Lcb3_scene_1/4'>LCB3</scene>, was developed using the same computational method as LCB1 and binds to the same region of the RBD, except in the opposite direction. Its <scene name='10/1076041/Lcb3xspikecorlabel_intxn/1'>binding interactions</scene> also show a large number of hydrogen bonds. However, LCB1 has more surface area and fits more precisely with the spike protein, giving it a slightly higher binding affinity (shown in the table to the right).[[Image:Kd_table.png|200px|right|'''Table 1'''. Dissociation constants (Kd) in nM for SARS-CoV-2 spike RBD binding to ACE2 and engineered minibinders, measured by biolayer interferometry to compare affinities for the spike protein.]] | ||
| - | =Significance= | + | ==Significance== |
The researchers demonstrated the advantages of using a de novo approach to design efficient miniprotein inhibitors that bind the SARS-CoV-2 spike protein with exceptionally high affinity. The dissociation constants (Kd) in nM for SARS-CoV-2 spike RBD binding to the different receptors are shown in the table to the right. The small, hyperstable miniproteins that were created using this approach (i.e., LCB1 and LCB3) are smaller, more stable, cheaper to produce, and better suited for delivery (e.g., nasal sprays) compared to traditional antibodies. By targeting the receptor-binding domain (RBD) and competing directly with ACE2, these inhibitors block viral entry into host cells, as they bind over 50 times more tightly than the ACE2 receptor. The final structures of LCB1 and LCB3 differed by around had very low deviation (~ 1.3-1.9 Å) from their computer models, demonstrating the accuracy of the computational tools used in predicting the three-dimensional folding of the miniproteins along with their specific interactions with the spike RBD. | The researchers demonstrated the advantages of using a de novo approach to design efficient miniprotein inhibitors that bind the SARS-CoV-2 spike protein with exceptionally high affinity. The dissociation constants (Kd) in nM for SARS-CoV-2 spike RBD binding to the different receptors are shown in the table to the right. The small, hyperstable miniproteins that were created using this approach (i.e., LCB1 and LCB3) are smaller, more stable, cheaper to produce, and better suited for delivery (e.g., nasal sprays) compared to traditional antibodies. By targeting the receptor-binding domain (RBD) and competing directly with ACE2, these inhibitors block viral entry into host cells, as they bind over 50 times more tightly than the ACE2 receptor. The final structures of LCB1 and LCB3 differed by around had very low deviation (~ 1.3-1.9 Å) from their computer models, demonstrating the accuracy of the computational tools used in predicting the three-dimensional folding of the miniproteins along with their specific interactions with the spike RBD. | ||
Revision as of 19:01, 28 April 2025
De Novo Miniprotein COVID-19 Therapeutic
| |||||||||||