Sandbox Reserved 1852
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
Overall, the Diels-Alderase stimulates improvement in synthetic laboratories and demonstrates early success in the now-prominent world of [https://www.nobelprize.org/prizes/chemistry/2024/press-release/ computational enzyme design.] | Overall, the Diels-Alderase stimulates improvement in synthetic laboratories and demonstrates early success in the now-prominent world of [https://www.nobelprize.org/prizes/chemistry/2024/press-release/ computational enzyme design.] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| Line 22: | Line 17: | ||
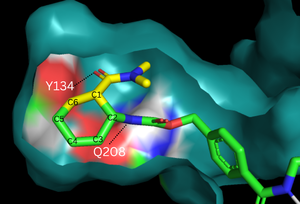
[[Image:Diels-AlderaseSurfaces.png|300px|left|thumb|Figure 2. Binding pocket and substrate. Shown is the binding pocket of the enzyme shown as surface, highlighting the electrostatics of the two catalytic residues, Tyr134 and Glu208. The ligand is color coded based on original structure: the dienophile is in yellow and the diene is in green. The reaction proceeds via attack of the C6 on the C5, shifting electron density to C2, which attacks C1.]] | [[Image:Diels-AlderaseSurfaces.png|300px|left|thumb|Figure 2. Binding pocket and substrate. Shown is the binding pocket of the enzyme shown as surface, highlighting the electrostatics of the two catalytic residues, Tyr134 and Glu208. The ligand is color coded based on original structure: the dienophile is in yellow and the diene is in green. The reaction proceeds via attack of the C6 on the C5, shifting electron density to C2, which attacks C1.]] | ||
====Scaffold==== | ====Scaffold==== | ||
| - | The original enzyme was found using Rosetta parameters that | + | The original enzyme scaffold was found using Rosetta parameters that searched for enzymes that contained catalytic Tyr and Glu residues and could coordinate two substrates highly specifically. Using the software, a catalog of 207 scaffolds was screened for potential active site orientations that could accommodate the substrates. 84 of the 207 were selected for testing, 50 were found to be soluble, and only 2, DA_20_00 and DA_42_00, had any enzymatic activity.<ref name="Siegel"/> Preliminary results favored <scene name='10/1075254/Squidscaffold/2'>DA_20_00</scene> and thus this [https://en.wikipedia.org/wiki/Beta-propeller beta-propeller] scaffold was chosen as the base for further experiments.<ref name="Siegel"/><ref name="Scharff">PMID:11435114</ref> The DA_20_00 protein scaffold is a 6-bladed beta-propeller of ''Loligo vulgalis,'' or the [https://en.wikipedia.org/wiki/European_squid European Squid]. The protein is relatively simple, with only one chain, one unit, 324 residues, and no extra ligands, metal ions, or small molecules bound. |
====Active Site==== | ====Active Site==== | ||
In the designed active site, <scene name='10/1075254/Active_site/6'>two catalytic residues</scene> stabilize the transition state of the Diels-Alder reaction. The Tyr134 acts as a <scene name='10/1075254/Y134_h_donation/3'>hydrogen bond donor</scene> to the oxygen on the dienophile [Fig. 2]. Q208 acts as a <scene name='10/1075254/208_bond_donor/3'>hydrogen bond acceptor</scene> to the nitrogen on the diene [Fig. 2]. These interactions help reduce the energetic gap between orbitals, allowing the reaction to proceed. The active site geometry also plays a large role in the binding of the substrates and how they react on a stereochemical level. By making small changes in the active site, changes can be made to the selectivity. | In the designed active site, <scene name='10/1075254/Active_site/6'>two catalytic residues</scene> stabilize the transition state of the Diels-Alder reaction. The Tyr134 acts as a <scene name='10/1075254/Y134_h_donation/3'>hydrogen bond donor</scene> to the oxygen on the dienophile [Fig. 2]. Q208 acts as a <scene name='10/1075254/208_bond_donor/3'>hydrogen bond acceptor</scene> to the nitrogen on the diene [Fig. 2]. These interactions help reduce the energetic gap between orbitals, allowing the reaction to proceed. The active site geometry also plays a large role in the binding of the substrates and how they react on a stereochemical level. By making small changes in the active site, changes can be made to the selectivity. | ||
====Helix Cap==== | ====Helix Cap==== | ||
In the evolution process, a 16-residue [https://proteopedia.org/wiki/index.php/Alpha_helix alpha-helix] <scene name='10/1075254/Alpha_helix_highlighted/1'>cap</scene> to the top of the binding site. The original hypothesis was that including a steric group near to the top of the active site would increase the binding affinity of the enzyme and improve the reaction kinetics. It was experimentally shown that he hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction,” decreasing the ''K<sub>m</sub>'' of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.<ref name="Eiben">PMID:22267011</ref> | In the evolution process, a 16-residue [https://proteopedia.org/wiki/index.php/Alpha_helix alpha-helix] <scene name='10/1075254/Alpha_helix_highlighted/1'>cap</scene> to the top of the binding site. The original hypothesis was that including a steric group near to the top of the active site would increase the binding affinity of the enzyme and improve the reaction kinetics. It was experimentally shown that he hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction,” decreasing the ''K<sub>m</sub>'' of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.<ref name="Eiben">PMID:22267011</ref> | ||
| - | |||
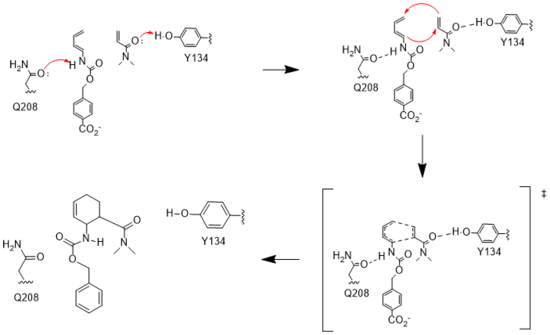
== Mechanism == | == Mechanism == | ||
| Line 55: | Line 49: | ||
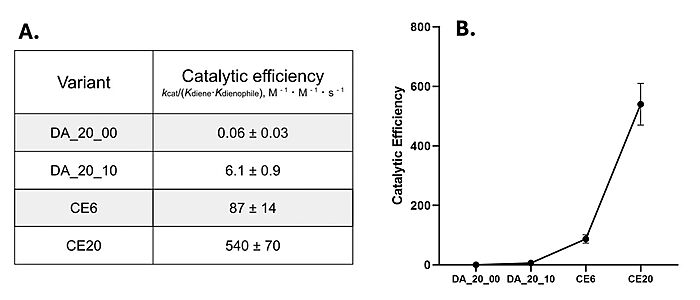
Classic [https://en.wikipedia.org/wiki/Michaelis%E2%80%93Menten_kinetics Michaelis-Menten kinetics]were determined for each generation of the enzyme. As the Diels-Alderase relies on a catalyzed interaction between both the diene and dienophile, a Michaelis binding constant (''K<sub>m</sub>'' value) was determined for each substrate separately before catalytic efficiency was calculated. The CE20 model of the enzyme is over 300-fold more efficient than the first enzyme model due to increasing active site specificity [Fig. 4].<ref name="Preiswerk"/> The authors did not publish comparisons to free reflux originally. | Classic [https://en.wikipedia.org/wiki/Michaelis%E2%80%93Menten_kinetics Michaelis-Menten kinetics]were determined for each generation of the enzyme. As the Diels-Alderase relies on a catalyzed interaction between both the diene and dienophile, a Michaelis binding constant (''K<sub>m</sub>'' value) was determined for each substrate separately before catalytic efficiency was calculated. The CE20 model of the enzyme is over 300-fold more efficient than the first enzyme model due to increasing active site specificity [Fig. 4].<ref name="Preiswerk"/> The authors did not publish comparisons to free reflux originally. | ||
| - | |||
| - | |||
Revision as of 20:41, 28 April 2025
Diels-Alderase
| |||||||||||