We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Carson Powers/Sandbox 1
From Proteopedia
< User:Carson Powers(Difference between revisions)
| Line 28: | Line 28: | ||
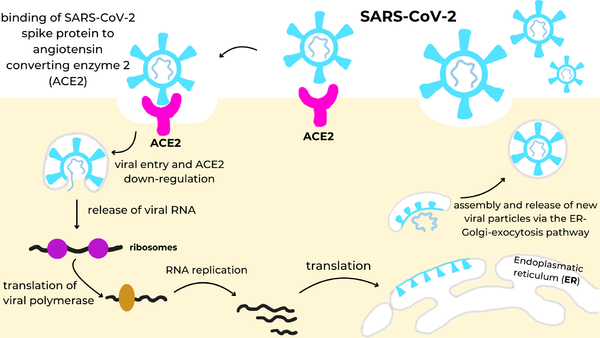
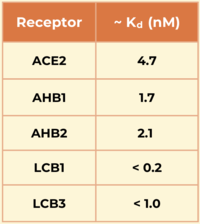
The researchers demonstrated the advantages of using a de novo approach to design efficient miniprotein inhibitors that bind the SARS-CoV-2 spike protein with exceptionally high affinity. The dissociation constants (Kd) in nM for SARS-CoV-2 spike protein binding to the different receptors are shown in the table to the right. The small, hyperstable miniproteins that were created using this approach (i.e., LCB1 and LCB3) are smaller, more stable, cheaper to produce, and better suited for delivery (e.g., nasal sprays) compared to traditional antibodies. By targeting the receptor-binding domain (RBD) and competing directly with ACE2, these inhibitors block viral entry into host cells, as they bind over 50 times more tightly than the ACE2 receptor. The final structures of LCB1 and LCB3 differed by around had very low deviation (~ 1.3-1.9 Å) from their computer models, demonstrating the accuracy of the computational tools used in predicting the three-dimensional folding of the miniproteins along with their specific interactions with the spike RBD. | The researchers demonstrated the advantages of using a de novo approach to design efficient miniprotein inhibitors that bind the SARS-CoV-2 spike protein with exceptionally high affinity. The dissociation constants (Kd) in nM for SARS-CoV-2 spike protein binding to the different receptors are shown in the table to the right. The small, hyperstable miniproteins that were created using this approach (i.e., LCB1 and LCB3) are smaller, more stable, cheaper to produce, and better suited for delivery (e.g., nasal sprays) compared to traditional antibodies. By targeting the receptor-binding domain (RBD) and competing directly with ACE2, these inhibitors block viral entry into host cells, as they bind over 50 times more tightly than the ACE2 receptor. The final structures of LCB1 and LCB3 differed by around had very low deviation (~ 1.3-1.9 Å) from their computer models, demonstrating the accuracy of the computational tools used in predicting the three-dimensional folding of the miniproteins along with their specific interactions with the spike RBD. | ||
| - | ==Therapeutic Relevance== | + | ==''Therapeutic Relevance''== |
The LCBs have a significantly higher affinity for the spike RBD and are smaller and more stable than traditional antibodies. Their reduced size allows these molecules to tightly pack more interactions in the active site as well as improves their ability to enter the respiratory system. Their hyperstability makes them able to withstand high and low temperatures, allowing them to be stored anywhere--contributing to their higher efficacy than most traditional antibodies.<ref name="Cao"/> However, further research is needed as they have not used the miniproteins in animal studies or human trials yet, and there is a small risk of immunogenicity. Another flaw in this method is it assumes no mutations occur. Since viruses mutate very quickly, by the time these therapeutics are distributed, they may be rendered useless due to a slight change in the spike protein structure. But overall, the development of LCB1 and LCB3 validates the reliability, efficiency, and accuracy of the computational design approach, demonstrating a quick and efficient strategy for producing therapeutics in response to emerging infectious diseases. | The LCBs have a significantly higher affinity for the spike RBD and are smaller and more stable than traditional antibodies. Their reduced size allows these molecules to tightly pack more interactions in the active site as well as improves their ability to enter the respiratory system. Their hyperstability makes them able to withstand high and low temperatures, allowing them to be stored anywhere--contributing to their higher efficacy than most traditional antibodies.<ref name="Cao"/> However, further research is needed as they have not used the miniproteins in animal studies or human trials yet, and there is a small risk of immunogenicity. Another flaw in this method is it assumes no mutations occur. Since viruses mutate very quickly, by the time these therapeutics are distributed, they may be rendered useless due to a slight change in the spike protein structure. But overall, the development of LCB1 and LCB3 validates the reliability, efficiency, and accuracy of the computational design approach, demonstrating a quick and efficient strategy for producing therapeutics in response to emerging infectious diseases. | ||
Current revision
De Novo Miniprotein COVID-19 Therapeutic
| |||||||||||