Journal:Acta Cryst F:S2053230X25007034
From Proteopedia
(Difference between revisions)

| Line 1: | Line 1: | ||
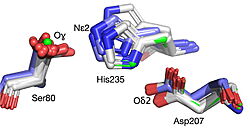
<StructureSection load='' size='450' side='right' scene='10/1087242/019_fig_2a_png/1' caption='The structure of HNL6V ([[8euo]]), an α/β-hydrolase fold enzyme. The catalytic triad (S80, H235, D207) orange sticks with the Oɣ of Ser80 in red at the center of the figure. The pink spheres show the Cɑ’s of the seven substitutions (T11G, E79H, C81L, H103V, D104A, G176S, K236M) the were engineered to convert hydroxynitrile lyase from rubber trees (HbHNL) to make it more structurally similar to SABP2, an esterase from tobacco that shares 44% sequence identity.'> | <StructureSection load='' size='450' side='right' scene='10/1087242/019_fig_2a_png/1' caption='The structure of HNL6V ([[8euo]]), an α/β-hydrolase fold enzyme. The catalytic triad (S80, H235, D207) orange sticks with the Oɣ of Ser80 in red at the center of the figure. The pink spheres show the Cɑ’s of the seven substitutions (T11G, E79H, C81L, H103V, D104A, G176S, K236M) the were engineered to convert hydroxynitrile lyase from rubber trees (HbHNL) to make it more structurally similar to SABP2, an esterase from tobacco that shares 44% sequence identity.'> | ||
===Crystal structure of a seven-substitution mutant of hydroxynitrile lyase from rubber tree=== | ===Crystal structure of a seven-substitution mutant of hydroxynitrile lyase from rubber tree=== | ||
| - | <big> | + | <big>Colin T. Pierce, Lauren R. Greenberg, Meghan E. Walsh, Ke Shi, Drenen J. Mageea, Hideki |
| + | Aihara, Wendy Gordon, Robert L. Evans III* and Romas J. Kazlauskas</big> <ref>doi: 10.1107/S2053230X25007034</ref> | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
Revision as of 20:39, 13 August 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.