Journal:Acta Cryst D:S2059798325007089
From Proteopedia
(Difference between revisions)

| Line 15: | Line 15: | ||
|- | |- | ||
|- | |- | ||
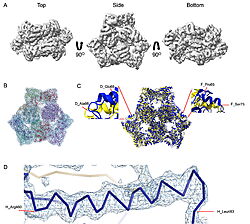
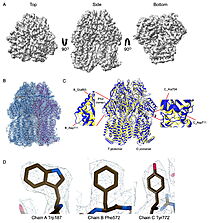
| - | | [[Image:022_Fig2.jpg|thumb|left|250px|Cryo-EM density map and model fitting of the hexameric ArnA complex. (a) ArnA hexamer reconstructed at 4.0 Å resolution, shown from three orientations: top, side, and bottom views. The map reveals the characteristic two-layered architecture of ArnA and clearly resolved secondary-structure elements. (b) Final atomic model of ArnA fitted into the cryo-EM density map. Each of the 12 subunits is displayed in a different color to illustrate the hexameric arrangement. (c) Structural alignment of the final cryo-EM model (blue) (PDB ID [[9y5h]]) with the reference crystal structure (yellow) (PDB ID [[6pih]]) using ChimeraX. The alignment yielded an rmsd of 1.2 Å. Enlarged views show local conformational deviations in loop regions: Glu69–Ala98 in chain D (left) and Pro65–Ser75 in chain F (right). (d) Close-up view showing the model-to-map fit for a peptide segment (Leu483–Arg460). The density mesh is contoured at 1. | + | | [[Image:022_Fig2.jpg|thumb|left|250px|Cryo-EM density map and model fitting of the hexameric ArnA complex. (a) ArnA hexamer reconstructed at 4.0 Å resolution, shown from three orientations: top, side, and bottom views. The map reveals the characteristic two-layered architecture of ArnA and clearly resolved secondary-structure elements. (b) Final atomic model of ArnA fitted into the cryo-EM density map. Each of the 12 subunits is displayed in a different color to illustrate the hexameric arrangement. (c) Structural alignment of the final cryo-EM model (blue) (PDB ID [[9y5h]]) with the reference crystal structure (yellow) (PDB ID [[6pih]]) using ChimeraX. The alignment yielded an rmsd of 1.2 Å. Enlarged views show local conformational deviations in loop regions: Glu69–Ala98 in chain D (left) and Pro65–Ser75 in chain F (right). (d) Close-up view showing the model-to-map fit for a peptide segment (Leu483–Arg460). The density mesh is contoured at 1.7σ , showing clear peptide backbone density and supporting accurate model placement.]] [[Image:022_Fig3.jpg|thumb|right|210px|Cryo-EM density map and model fitting of the AcrB trimer. (a) AcrB reconstructed at 2.92 Å resolution, shown from top, side, and bottom views. It reveals secondary-structure elements and transmembrane helices, consistent with the known trimeric architecture. (b) Final atomic model of AcrB fitted into the cryo-EM density map. Each protomer is shown in a different color to highlight the asymmetric trimer organization. (c) Structural alignment of the final cryo-EM model (blue)(PDB-ID [[9v5r]]) with the reference crystal structure , (yellow) (PDB ID [[7rr7]]), performed using ChimeraX. The rmsd between the structures is 1.2 Å. Enlarged views highlight conformational deviations, including E693–D711 in protomer B (left) and A704–D711 in protomer C (right), located within the PN2 subdomain. (d) Close-up view showing the density fit for side chains (W187, chain A, F572, chain B, W772. chain C). The mesh is contoured at 1.6σ in Coot, showing well-defined density for aromatic residues.]] |
|} | |} | ||
Revision as of 08:31, 18 September 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.