Journal:Acta Cryst D:S2059798325007089
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
When cells build and maintain their membranes, they need a balance of protein insertion, folding, and degradation. In ''Escherichia coli'', this process is hypothetic to involve the AAA+ protease FtsH, the insertase YidC, and the regulatory HflKC complex. Their interaction had not been shown at structural level. To test this idea, we used single-particle cryo-electron microscopy on detergent-solubilized samples enriched for these proteins. | When cells build and maintain their membranes, they need a balance of protein insertion, folding, and degradation. In ''Escherichia coli'', this process is hypothetic to involve the AAA+ protease FtsH, the insertase YidC, and the regulatory HflKC complex. Their interaction had not been shown at structural level. To test this idea, we used single-particle cryo-electron microscopy on detergent-solubilized samples enriched for these proteins. | ||
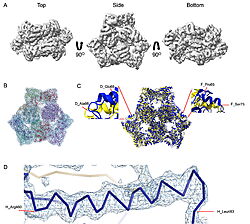
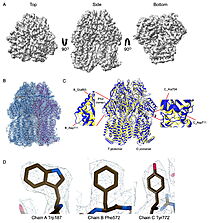
| - | The results were unexpected. Instead of clear views of an FtsH–HflKC–YidC assembly, the datasets revealed <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA</scene>, an enzyme linked to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the multidrug efflux transporter of the AcrAB–TolC system. Both proteins are known to appear during affinity purification, but their repeated presence across different methods and even in membrane fractions suggests that their recovery was not purely accidental. ArnA, usually described as cytoplasmic, was consistently found in membrane-enriched samples, while AcrB is a well-established membrane protein. We also observed class averages resembling GroEL and cytochrome bo3 oxidase. | + | The results were unexpected. Instead of clear views of an FtsH–HflKC–YidC assembly, the datasets revealed <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA</scene>, an enzyme linked to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the multidrug efflux transporter of the AcrAB–TolC system. Structural alignment of the final cryo-EM model (PDB ID [[9v5r]] (blue)) with the reference crystal structure (PDB ID [[7rr7]], yellow) shows that they are remarkably similar with an RMSD between them, for CA atoms, of 2.0Å. |
| + | |||
| + | |||
| + | Both proteins are known to appear during affinity purification, but their repeated presence across different methods and even in membrane fractions suggests that their recovery was not purely accidental. ArnA, usually described as cytoplasmic, was consistently found in membrane-enriched samples, while AcrB is a well-established membrane protein. We also observed class averages resembling GroEL and cytochrome bo3 oxidase. | ||
These findings show that cryo-EM can capture not only the intended targets but also unexpected complexes that are well resolved and may have physiological importance. While only partial densities of the FtsH AAA+ domain were visible and no stable FtsH–YidC–HflKC complex could be reconstructed, the high-quality ArnA and AcrB structures provide fresh insights into bacterial survival strategies, from antibiotic resistance to drug efflux. More broadly, this study illustrates how structural biology can reveal unplanned discoveries that enrich our understanding of cell biology and the challenges of protein purification. | These findings show that cryo-EM can capture not only the intended targets but also unexpected complexes that are well resolved and may have physiological importance. While only partial densities of the FtsH AAA+ domain were visible and no stable FtsH–YidC–HflKC complex could be reconstructed, the high-quality ArnA and AcrB structures provide fresh insights into bacterial survival strategies, from antibiotic resistance to drug efflux. More broadly, this study illustrates how structural biology can reveal unplanned discoveries that enrich our understanding of cell biology and the challenges of protein purification. | ||
Revision as of 15:55, 19 September 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.