Journal:Acta Cryst D:S2059798325007089
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
When cells build and maintain their membranes, they need a balance of protein insertion, folding, and degradation. In ''Escherichia coli'', this process is hypothetic to involve the AAA+ protease FtsH, the insertase YidC, and the regulatory HflKC complex. Their interaction had not been shown at structural level. To test this idea, we used single-particle cryo-electron microscopy on detergent-solubilized samples enriched for these proteins. | When cells build and maintain their membranes, they need a balance of protein insertion, folding, and degradation. In ''Escherichia coli'', this process is hypothetic to involve the AAA+ protease FtsH, the insertase YidC, and the regulatory HflKC complex. Their interaction had not been shown at structural level. To test this idea, we used single-particle cryo-electron microscopy on detergent-solubilized samples enriched for these proteins. | ||
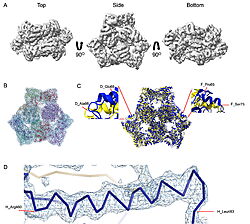
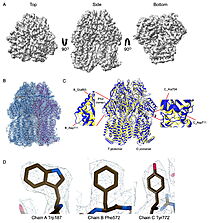
| - | The results were unexpected. Instead of clear views of an FtsH–HflKC–YidC assembly, the datasets revealed <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA</scene>, an enzyme linked to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the multidrug efflux transporter of the AcrAB–TolC system. Structural alignment of the final cryo-EM model (PDB ID [[9v5r]] (blue)) with the reference crystal structure (PDB ID [[7rr7]], (yellow)) shows that they are remarkably similar, with an <scene name='10/1089031/022_aligned_9v5r_on_7rr7/1'>RMSD between them, for CA atoms, of 2.0Å | + | The results were unexpected. Instead of clear views of an FtsH–HflKC–YidC assembly, the datasets revealed <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA</scene>, an enzyme linked to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the multidrug efflux transporter of the AcrAB–TolC system. Structural alignment of the final cryo-EM model (PDB ID [[9v5r]] (blue)) with the reference crystal structure (PDB ID [[7rr7]], (yellow)) shows that they are remarkably similar, with an <scene name='10/1089031/022_aligned_9v5r_on_7rr7/1'>superposion of Cryo-EM vs crystal structure for AcrB</scene> with an RMSD between them, for CA atoms, of 2.0Å. |
Revision as of 16:04, 19 September 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.