Journal:Acta Cryst D:S2059798325007089
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
Cells rely on finely tuned machinery to build and maintain their membranes, balancing the insertion, folding, and removal of proteins. In ''E. coli'', this process is thought to involve the protease FtsH, the insertase YidC, and the regulatory HflKC complex. But their partnership had never been seen directly. To investigate, researchers turned to single-particle cryo-electron microscopy on detergent-solubilized samples enriched in these proteins. | Cells rely on finely tuned machinery to build and maintain their membranes, balancing the insertion, folding, and removal of proteins. In ''E. coli'', this process is thought to involve the protease FtsH, the insertase YidC, and the regulatory HflKC complex. But their partnership had never been seen directly. To investigate, researchers turned to single-particle cryo-electron microscopy on detergent-solubilized samples enriched in these proteins. | ||
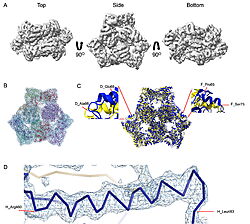
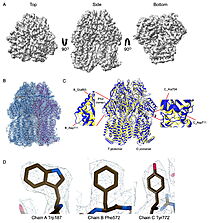
| - | What they found was surprising. Instead of the expected three-part assembly, the cryo-EM maps revealed exquisite structures of two other players: <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA/scene>, an enzyme tied to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the major drug efflux transporter. Structural alignment of the final cryo-EM model (PDB ID [[9v5r]] (blue)) with the reference crystal structure (PDB ID [[7rr7]], (yellow)) shows that they are remarkably similar, with an <scene name='10/1089031/022_aligned_9v5r_on_7rr7/1'>superposion of Cryo-EM vs crystal structure for AcrB</scene> with an RMSD between them, for CA atoms, of 2.0Å and <scene name='10/1089031/022_aligned_9v5r_on_7rr7/3'>animation</scene>. | + | What they found was surprising. Instead of the expected three-part assembly, the cryo-EM maps revealed exquisite structures of two other players: <scene name='10/1089031/022_fig_arna_hexamer/1'>high-resolution structures of ArnA</scene>, an enzyme tied to polymyxin resistance, and <scene name='10/1089031/022_fig_arcb_trimer/1'>AcrB</scene>, the major drug efflux transporter. Structural alignment of the final cryo-EM model (PDB ID [[9v5r]] (blue)) with the reference crystal structure (PDB ID [[7rr7]], (yellow)) shows that they are remarkably similar, with an <scene name='10/1089031/022_aligned_9v5r_on_7rr7/1'>superposion of Cryo-EM vs crystal structure for AcrB</scene> with an RMSD between them, for CA atoms, of 2.0Å and <scene name='10/1089031/022_aligned_9v5r_on_7rr7/3'>animation</scene>. |
Both proteins are known to appear during affinity purification, but their repeated presence across different methods and even in membrane fractions suggests that their recovery was not purely accidental. ArnA, usually described as cytoplasmic, was consistently found in membrane-enriched samples, while AcrB is a well-established membrane protein. We also observed class averages resembling GroEL and cytochrome bo3 oxidase. | Both proteins are known to appear during affinity purification, but their repeated presence across different methods and even in membrane fractions suggests that their recovery was not purely accidental. ArnA, usually described as cytoplasmic, was consistently found in membrane-enriched samples, while AcrB is a well-established membrane protein. We also observed class averages resembling GroEL and cytochrome bo3 oxidase. | ||
Revision as of 17:21, 2 October 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.