We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
HOAT1
From Proteopedia
(Difference between revisions)
| Line 149: | Line 149: | ||
| - | === | + | ===Mechanistic Insights into hOAT1 Function and Inhibition=== |
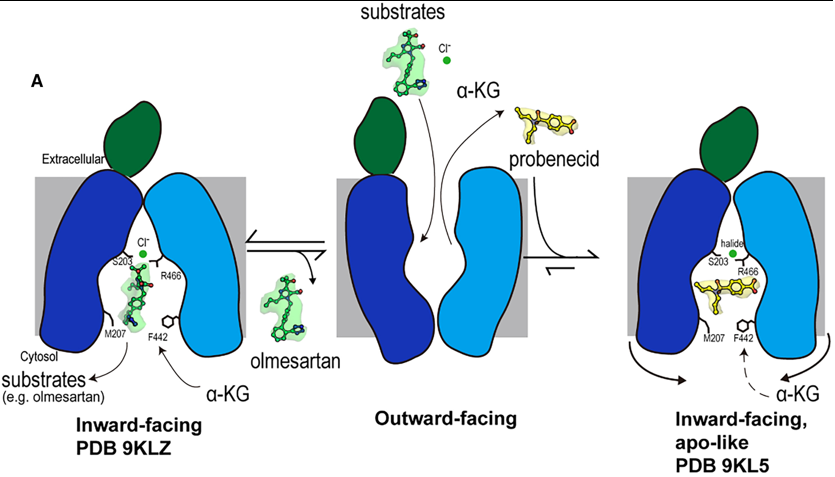
[[Image:HOAT1mechanism.png | frame |300px| upright= 1.5 |none | alt= | Fig 2. Mechanism of olmesartan binding and conformational inhibition by probenecid. A) When the transporter is in its outward-facing conformation, substrates or inhibitors enter the central binding pocket and undergo structural rearrangement to | [[Image:HOAT1mechanism.png | frame |300px| upright= 1.5 |none | alt= | Fig 2. Mechanism of olmesartan binding and conformational inhibition by probenecid. A) When the transporter is in its outward-facing conformation, substrates or inhibitors enter the central binding pocket and undergo structural rearrangement to | ||
| Line 156: | Line 156: | ||
conformation change for inhibition (apo-like conformation).]] | conformation change for inhibition (apo-like conformation).]] | ||
| - | ''' | + | '''1. A Dual-Mechanism for Potent Inhibition by Probenecid''' |
| - | + | The study reveals that the classic inhibitor probenecid employs a sophisticated, dual-mechanism to arrest OAT1 function, moving beyond simple competition. | |
| - | ''' | + | '''Direct Competition:''' Probenecid occupies the central binding pocket, and its interaction with K382 in Site 1 directly competes with the binding of the counter-substrate α-ketoglutarate (α-KG). This disrupts the exchange cycle that drives substrate transport. |
| - | ''' | + | '''Conformational Arrest:''' More significantly, probenecid binding induces subtle conformational changes in the cytoplasmic ends of transmembrane helices (TM5, TM8, TM10, TM11). This leads to a constriction of the cytosolic opening, completely blocking one access path (Path B) and narrowing the other (Path A). This physically prevents substrates from entering or exiting the binding site from the cytoplasm, effectively "locking" the transporter in an inactive, inward-facing state. This mechanism is reminiscent of inhibition seen in other transporters like hURAT1, suggesting it may be a general strategy for effective transport arrest. |
| - | + | '''2. Structural Basis for Species-Specific Drug Transport''' | |
| - | + | A major advancement of this work is the structural explanation for long-observed differences in drug handling between human OAT1 and animal orthologs. | |
| - | + | '''The Critical Role of S203:''' The residue S203 in hOAT1 (which is an alanine, A203, in rat OAT1) is identified as a key species-specific determinant. It does not contact substrates like olmesartan directly. Instead, its hydroxyl group is crucial for coordinating a chloride ion along with residues Y230 and R466. | |
| - | + | '''Chloride Coordination Enhances Substrate Affinity:''' This chloride coordination network stabilizes the binding of certain substrates. Functional data confirms that the S203A mutation drastically reduces olmesartan affinity specifically in the presence of chloride. This explains why drugs like olmesartan and tenofovir show different transport kinetics between species; the human transporter, with its S203, has a enhanced, chloride-dependent mechanism for high-affinity binding that the rat ortholog lacks. | |
| - | '''4. Substrate Release:''' The inward-facing conformation with its open paths (Path A and Path B) allows the substrate to dissociate into the cytoplasm. The transporter then likely resets to the outward-facing state, driven by the exchange with intracellular α-ketoglutarate (α-KG). | ||
| - | '''Inhibition Mechanism (e.g., Probenecid)''' | ||
| - | The inhibitor probenecid exploits the transport cycle but arrests it through a dual mechanism: | ||
| - | |||
| - | '''1. Binding and Competition:''' | ||
| - | |||
| - | *Probenecid enters the binding pocket from the extracellular side and binds in the inward-facing conformation. | ||
| - | |||
| - | *It occupies Site 3 and partially extends into Site 1. In Site 1, it directly competes with the counter-substrate α-KG by forming a key hydrogen bond with K382, a residue critical for α-KG binding. | ||
| - | |||
| - | '''2. Conformational Arrest and Cytoplasmic Blockade:''' | ||
| - | |||
| - | *This is the primary inhibitory mechanism. Probenecid binding induces subtle but critical conformational changes in the cytoplasmic regions of TM5, TM8, TM10, and TM11. | ||
| - | |||
| - | *These helices shift inward, causing a constriction of the entire cytoplasmic opening of the binding pocket. | ||
| - | |||
| - | *This constriction completely blocks Path B and severely narrows Path A. | ||
| - | |||
| - | *By physically obstructing these cytosolic paths, probenecid achieves two things: | ||
| - | |||
| - | :*It prevents intracellular substrates from entering the binding pocket. | ||
| - | |||
| - | :*It traps the transporter in a locked, inward-facing, apo-like conformation, preventing the conformational changes needed to complete the transport cycle. | ||
===Author=== | ===Author=== | ||
Revision as of 16:38, 30 November 2025
Interactive 3D Complement in Proteopedia
|
| |
|
Cryo-EM structures of human OAT1 reveal drug binding and inhibition mechanisms[1]. | |
|
Cell Volume 33, Issue 11, P1856-1866.E5, November 06, 2025 |
Structure Tour
| |||||||||||